MBD Enriched Oyster DNA Pico Methyl-Seq Library Prep
Using the Zymo Pico Methyl Seq Kit on the MBD enriched Eastern Oyster DNA from samples from Rebecca Stevick
Dilution of MBD enriched DNA samples 2020-01-08
While we haven’t tested this kit on oyster DNA, my plan was to start with 1ng input DNA into the kit like what has worked for coral DNA. First the samples had to be diluted 1:2 to avoid pipetting less than a µl of DNA
| Sample | volume DNA | volume ultra pure water |
|---|---|---|
| 1 | 5 | 10 |
| 2 | 5 | 10 |
| 3 | 5 | 10 |
| 4 | 5 | 10 |
| 5 | 5 | 10 |
| 6 | 5 | 10 |
| 7 | 5 | 10 |
| 8 | 5 | 10 |
| 9 | 5 | 10 |
| 10 | 5 | 10 |
| 11 | 5 | 10 |
| 12 | 5 | 10 |
Sample Prep for PMS
Samples need to all include 1ng of sample, .05ng spike-in, and water up to 20µl
| sample | volume DNA | volume diluted spike | volume ultra pure water to 20µl |
|---|---|---|---|
| 1 | 1.3µl | 2µl | 16.7µl |
| 2 | 1.2µl | 2µl | 16.8µl |
| 3 | 1.7µl | 2µl | 16.3µl |
| 4 | 1.3µl | 2µl | 16.7µl |
| 5 | 1.7µl | 2µl | 16.3µl |
| 6 | 1.3µl | 2µl | 16.7µl |
| 7 | 2.1µl | 2µl | 15.9µl |
| 8 | 3.6µl | 2µl | 14.4µl |
| 9 | 1.5µl | 2µl | 16.5µl |
| 10 | 1.7µl | 2µl | 16.3µl |
| 11 | 2.2µl | 2µl | 15.8µl |
| 12 | 1.5µl | 2µl | 16.5µl |
Bisulfite Conversion
- Added 130µl lightning conversion reagent to each sample in at PCR tube
- Put tubes in the thermocycler Pico bisulfite conversion program
- After program sample tubes were put in the 4 degree fridge overnight
Next Day 2020-01-09 Cleanup
- Made 12 spin column, one for each sample
- Added 600µl M-binding buffer each to 12 spin columns
- Added 150µl of the BS reaction (all) to each individual tube
- Invert columns to mix
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 100µl M-Wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl L-desulfonation buffer to each column
- Let them sit for 15 minutes
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 2 minutes and discarded flowthrough
- Added 8µl warmed (56C) DNA elution buffer to each column and let sit for 5 minutes in new 1.5mL tubes to collect
- Centrifuged columns at 12,000 rcf for 30 seconds
Amplification with PrepAmp Primers
- Made Priming master mix on ice:
- 2µl 5X PrepAmp buffer * 12.5 = 25µl
- 1µl PrepAmp Primers (40µM) * 12.5 = 12.5µl
- Made new PCR tubes with 3µl of Priming MM and 7µl of bisulfite treated DNA
- Kept those on ice
- Made PrepAmp Mix on ice:
- 1µl 5X PrepAmp buffer * 12.5 = 12.5µl
- 3.75µl PrepAmp PreMix * 12.5 = 46.9µl
- 0.3µl PrepAmp polymerase * 12.5 = 3.75µl
- Made slightly diluted PrepAmp polymerase to avoid adding less than .5µl:
- 3.75µl PrepAmp polymerase
- 2.5µl elution buffer
- Set thermocylcer program with lid temp restricted to 25 degrees C and place samples inside and run the pico priming program on the thermocycler:
- 98 for 2 minutes
- 8 degrees for 1 minute
- 8 degree hold
- During hold vortex, spin tubes down, add 5.05µl PrepAmp Mix to each tube, vortex, spin down, and place back in thermocycler
- 8 degrees for 4 minutes
- 16 degrees for 1 minute with 3% ramp rate
- 22 degrees for 1 minute with 3% ramp rate
- 28 degrees for 1 minute with 3% ramp rate
- 36 degrees for 1 minute with 3% ramp rate
- 36.5 degrees for 1 minute with 3% ramp rate
- 37 degrees for 8 minutes
- repeat back from the first step one time through again
- During hold, vortex, spin tubes down, tried to add 0.3µl of slightly diluted PrepAmp polymerase, vortex, spin down, and place back into thermocycler
Cleanup with DNA Clean and Concentrator Columns (DCC)
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 107.45µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (15.35µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf for 2 minutes, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 5 minutes
- Centrifuged 12,000 rcf 30 seconds
First Amplification
- Made 1st Amp master mix:
- 12.5µl 2X Library Amp Mix * 12.5 = 156.25µl
- 1µl Library Amp Primer(10µM) * 12.5 = 12.5µl
- 12.5µl 2X Library Amp Mix * 12.5 = 156.25µl
- Added 13.5µl MM to new PCR tubes
- Added 11.5µl of cleaned and concentrated DNA sample to the appropriate new PCR tube
- Vortexed, spun down, and placed in thermocycler program 1st Pico Methyl Amp program 8 cycles for the 1ng input
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 2 minutes, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12.5µl warmed elution buffer to each column directly
- Incubated 5 minutes
- Centrifuged 12,000 rcf 30 seconds
Amplification with Index Primers
-
In PCR tubes combined the following:
sample volume DNA volume LibAmp MM volume i5 primer volume i7 primer 1 12µl 14µl 1µl 1 1µl 1 2 12µl 14µl 1µl 2 1µl 2 3 12µl 14µl 1µl 3 1µl 3 4 12µl 14µl 1µl 4 1µl 4 5 12µl 14µl 1µl 5 1µl 5 6 12µl 14µl 1µl 6 1µl 6 7 12µl 14µl 1µl 7 1µl 7 8 12µl 14µl 1µl 8 1µl 8 9 12µl 14µl 1µl 9 1µl 9 10 12µl 14µl 1µl 10 1µl 10 11 12µl 14µl 1µl 11 1µl 11 12 12µl 14µl 1µl 12 1µl 12 -
Vortexed, spun down, and placed in thermocycler program 2nd Pico Methyl Amp program (10 cycles)
1X Bead Cleanup
- Allowed beads to come to room temp after resuspending
- Made fresh 80% EtOH
- Added equal volume of beads to each sample: 26µl
- Pipetted to mix
- Incubated on shaker 15 minutes
- Followed normal bead cleanup procedure
- Resuspended and eluted beads in 15µl elution buffer
Broad Range Qubit
| Standard 1 | Standard 2 | Sample | Average DNA ng/µl |
|---|---|---|---|
| 168 RFU | 22757 RFU | 1 | 2.01 |
| - | - | 2 | 2 |
| - | - | 3 | 3.4 |
| - | - | 4 | 2 |
| - | - | 5 | 2.01 |
| - | - | 6 | 2.42 |
| - | - | 7 | 2.34 |
| - | - | 8 | 2.03 |
| - | - | 9 | 2.12 |
| - | - | 10 | 4.32 |
| - | - | 11 | 2.48 |
| - | - | 12 | 2 |
Not great concentrations, and we are out of DNA screen tapes. But it probably worked fine and just needs to be amplified further.
4 Cycle PCR and Cleanup
- Split up libraries by two to do two amplifications for each
- Made 24 PCR tubes with 7µl of sample in each
- Made a master mix of LibraryAmp Mix and primers and water for each sample pair
| sample mix | volume LibAmp MM | volume i5 primer | volume i7 primer | volume water |
|---|---|---|---|---|
| 1 | 24µl | 2µl 1 | 2µl 1 | 8µl |
| 2 | 24µl | 2µl 2 | 2µl 2 | 8µl |
| 3 | 24µl | 2µl 3 | 2µl 3 | 8µl |
| 4 | 24µl | 2µl 4 | 2µl 4 | 8µl |
| 5 | 24µl | 2µl 5 | 2µl 5 | 8µl |
| 6 | 24µl | 2µl 6 | 2µl 6 | 8µl |
| 7 | 24µl | 2µl 7 | 2µl 7 | 8µl |
| 8 | 24µl | 2µl 8 | 2µl 8 | 8µl |
| 9 | 24µl | 2µl 9 | 2µl 9 | 8µl |
| 10 | 24µl | 2µl 10 | 2µl 10 | 8µl |
| 11 | 24µl | 2µl 11 | 2µl 11 | 8µl |
| 12 | 24µl | 2µl 12 | 2µl 12 | 8µl |
- Added 18µl of the appropriate master mix to each tube
- Placed in thermocycler 2nd Amp program 4 cycles
1X Bead Cleanup
- Allowed beads to come to room temp after resuspending
- Made fresh 80% EtOH
- Duplicate amplifications were pooled back together for a total of 50µl
- Added equal volume of beads to each sample: 50µl
- Pipetted to mix
- Incubated on shaker 15 minutes
- Followed normal bead cleanup procedure
- Resuspended and eluted beads in 14µl elution buffer
Broad Range Qubit
| Standard 1 | Standard 2 | Sample | Average DNA ng/µl |
|---|---|---|---|
| 154 RFU | 22290 RFU | 1 | 18 |
| - | - | 2 | 18.7 |
| - | - | 3 | 30.8 |
| - | - | 4 | 19 |
| - | - | 5 | 13 |
| - | - | 6 | 26.3 |
| - | - | 7 | 23.8 |
| - | - | 8 | 22.5 |
| - | - | 9 | 20.2 |
| - | - | 10 | 39.2 |
| - | - | 11 | 26.8 |
| - | - | 12 | 14 |
D5000 TapeStation
Size distributions of libraries, and one example trace:
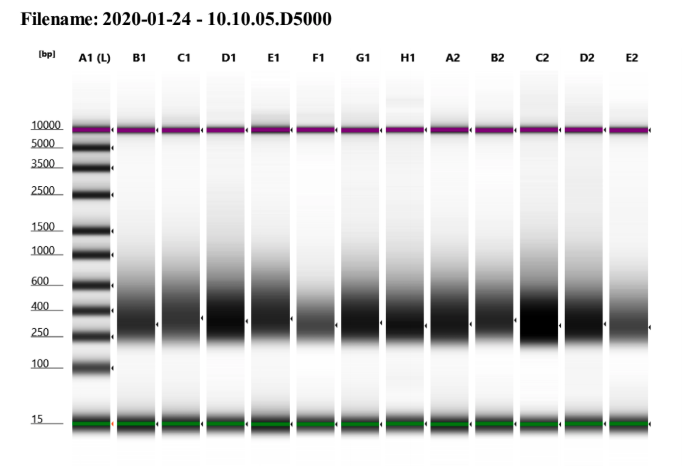
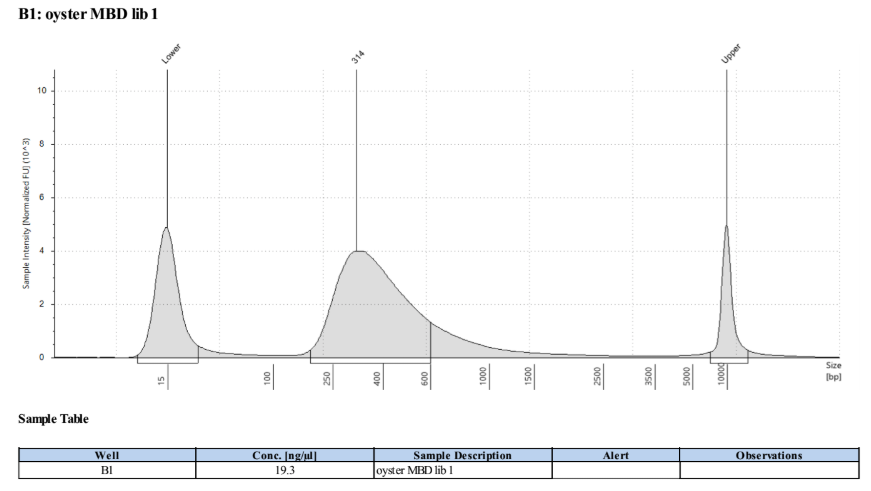
Final Sample Information
| Sample ID | Data_Name | Sample_Num | Sample_Name | Bucket | Treatment | Library ng/ul | i5 Index | Index i5 Sequence | i7 Index | Index i7 Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CON7_1 | 89 | BC.7.1 | 7 | Control | 18 | 1 | AGCGCTAG | 1 | CCGCGGTT |
| 2 | CON7_2 | 90 | BC.7.2 | 7 | Control | 18.7 | 2 | GATATCGA | 2 | TTATAACC |
| 3 | CON7_3 | 91 | BC.7.3 | 7 | Control | 30.8 | 3 | CGCAGACG | 3 | GGACTTGG |
| 4 | CON9_1 | 95 | BC.9.1 | 9 | Control | 19 | 4 | TATGAGTA | 4 | AAGTCCAA |
| 5 | CON9_2 | 96 | BC.9.2 | 9 | Control | 13 | 5 | AGGTGCGT | 5 | ATCCACTG |
| 6 | CON9_3 | 97 | BC.9.3 | 9 | Control | 26.3 | 6 | GAACATAC | 6 | GCTTGTCA |
| 7 | ENR11_1 | 101 | BC.11.1 | 11 | Enriched | 23.8 | 7 | ACATAGCG | 7 | CAAGCTAG |
| 8 | ENR11_2 | 102 | BC.11.2 | 11 | Enriched | 22.5 | 8 | GTGCGATA | 8 | TGGATCGA |
| 9 | ENR11_3 | 103 | BC.11.3 | 11 | Enriched | 20.2 | 9 | CCAACAGA | 9 | AGTTCAGG |
| 10 | ENR12_1 | 104 | BC.12.1 | 12 | Enriched | 39.2 | 10 | TTGGTGAG | 10 | GACCTGAA |
| 11 | ENR12_2 | 105 | BC.12.2 | 12 | Enriched | 26.8 | 11 | CGCGGTTC | 11 | TCTCTACT |
| 12 | ENR12_3 | 106 | BC.12.3 | 12 | Enriched | 14 | 12 | TATAACCT | 12 | CTCTCGTC |