Porites astrodies RNA Extraction with Purelink Kit
Using RNA Purelink Kit to Extract RNA from 3 Porites asteroides Samples Following Protocol from Federica
In this Extraction I used: Purelink RNA mini Kit, Trizol Reagent, 1-Bromo-3-cholropropane, and a protocol from Federica
Goal: Extract high quality RNA out of these samples to send to Israel
Result: No RNA extracted :(
Major Take Aways: Not sure what went wrong, my main guess is that it did not homogenize enough to break open the cells
Note: Protocol is done in the hood as Trizol and 1-Bromo-3-cholropropane are toxic and corrosive. PPE of lab coat, mask, eye glasses, and gloves were worn at all times. Solid and liquid wastes are hazardous.
Sample Prep
- Samples:
- R13 A2 201907 Molec
- R5 A2 201907 Molec
- R14 A2 201907 Molec
- Sterilized clippers and forceps with 10% bleach, DI water, 70% ETOH, and RNase Away
- Clipped fragment on dry ice at RNA bench, brought dry ice cooler to the hood, added 550ul trizol to a glass bead tube, and used the forceps to put the chip in the bead tube
- Repeated sterilization and sample addition inside the hood for each sample
- Tried to chip ~1cm2 of the flash frozen sample (what Federica uses)
- Sample R14 wouldn’t chip (too thick) so I just scraped from the tissue until there was a good amount to add
- Sample R13 in bead tube:
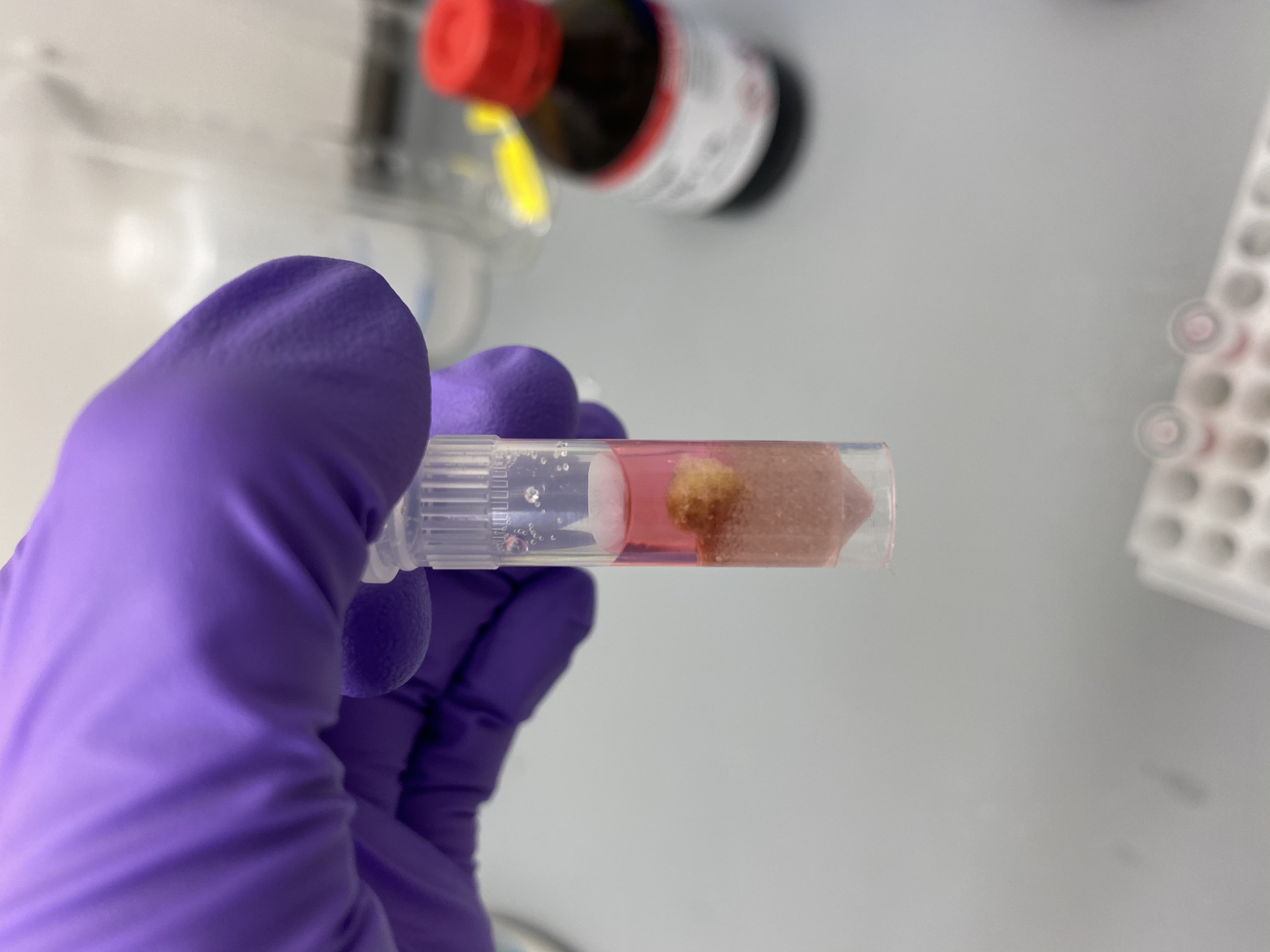
- Sample R5 in bead tube:
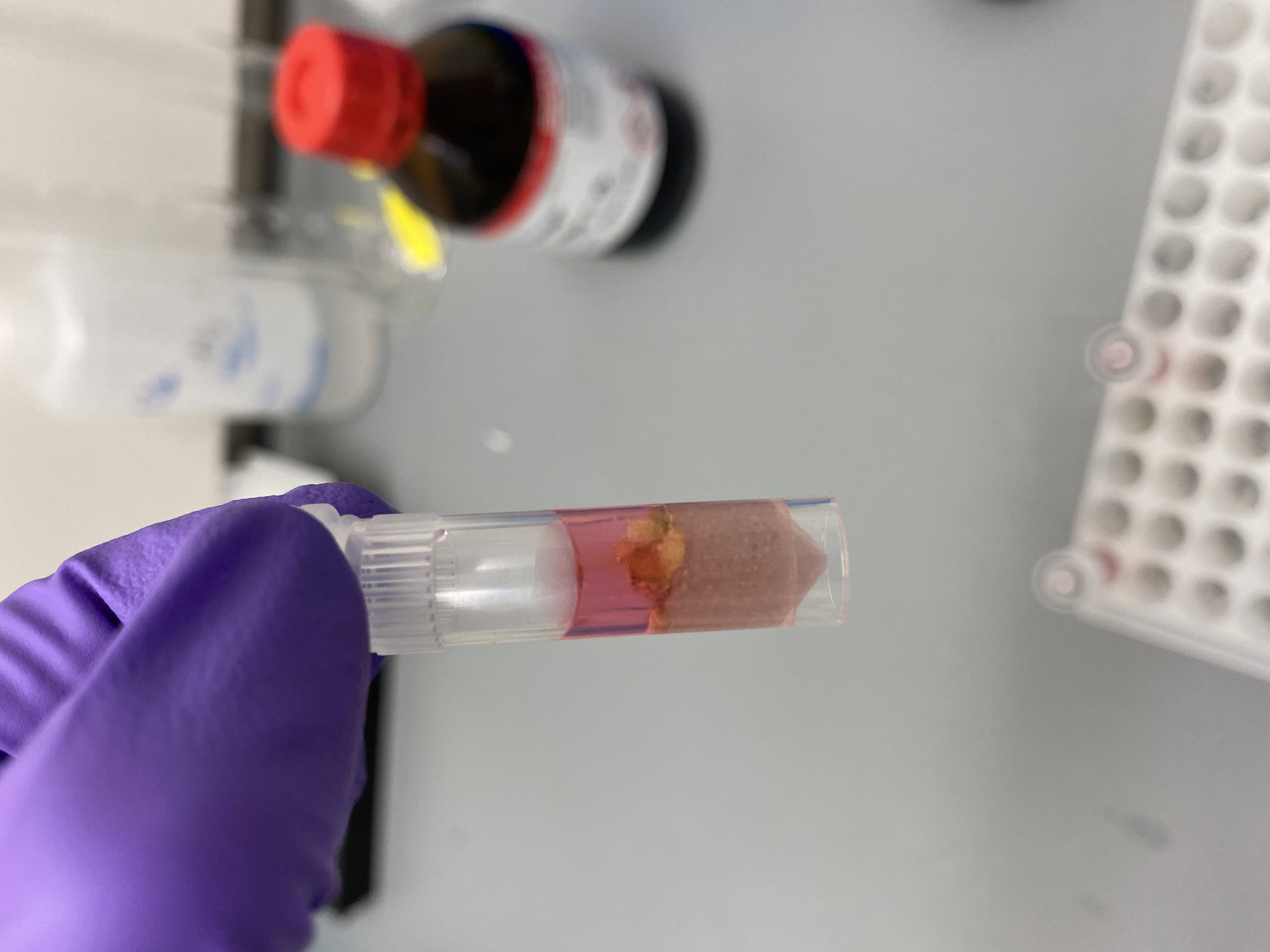
- Sample R14 in bead tube:
- Put bead tubes in the tissuelyser 2 minutes at 15Hz
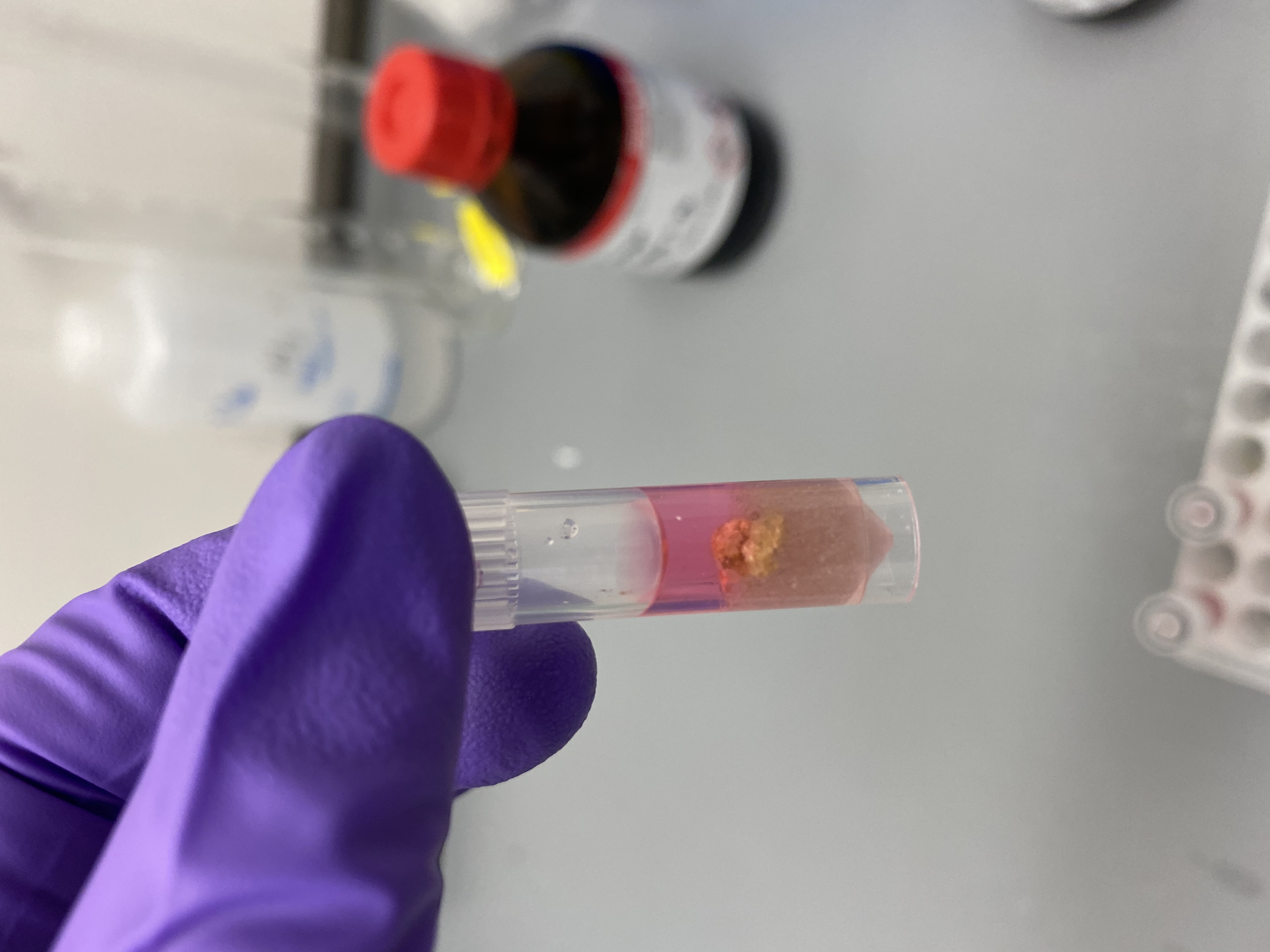
- R13 after tissuelyser, hard to see but in box looked like the whole tissue chip, tissue seems still very present on skeleton:
- R5 after tissuelyser, also doesn’t seem very broken up:
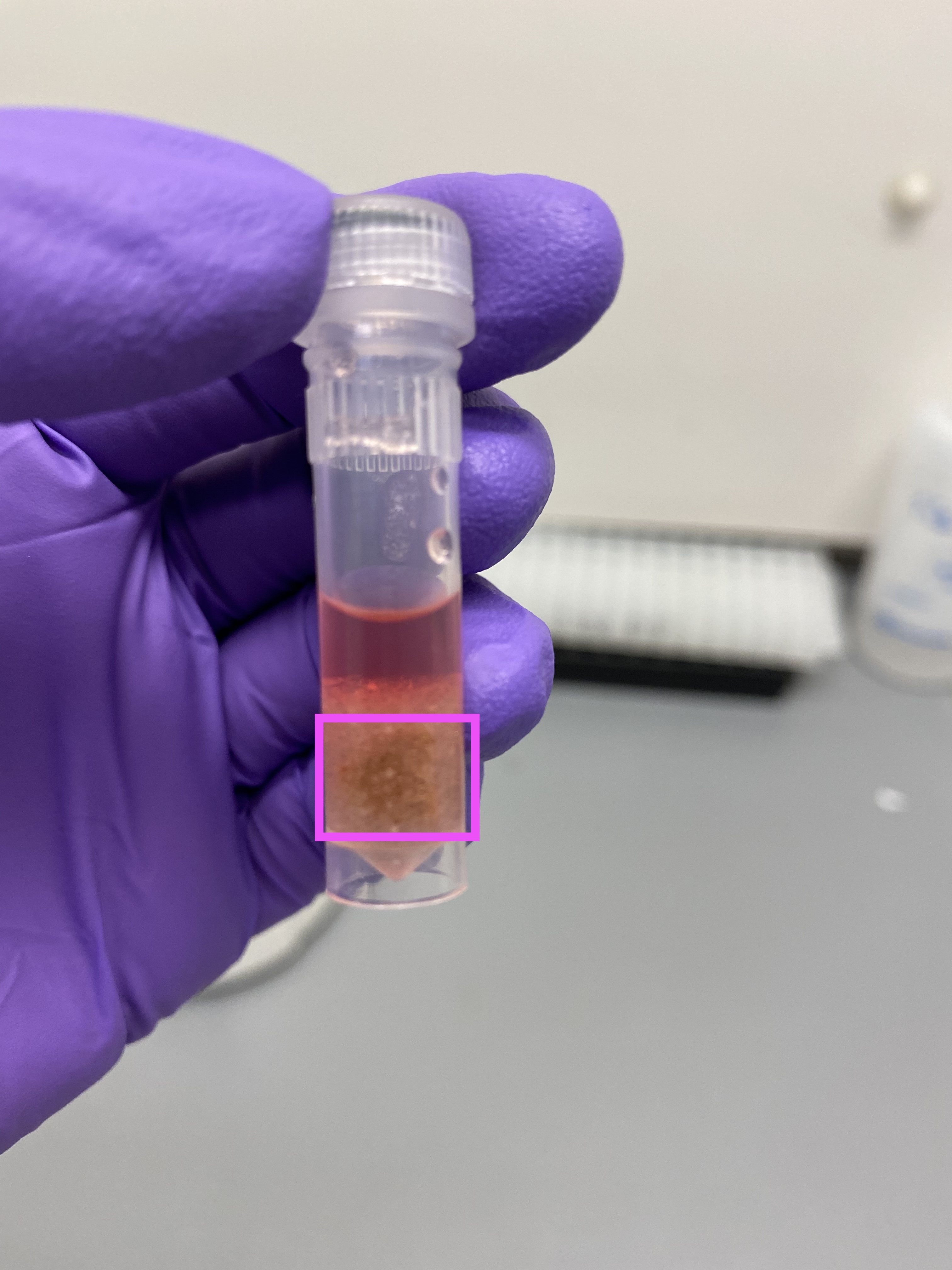
- R14 after tissuelyser, trizol looks noticeably browner thank other samples:
Extraction
- Removed ~350ul from bead tubes and put into new 1.5mL tubes
- Added 450ul of Trizol to each new 1.5mL tube, vortexed tubes briefly and spun down
- R13
- R5
- R14
- Incubated tubes at room temp for 5 minutes
- Added 100ul of 1-Bromo-3-cholropropane and vortexed tubes for 15 seconds each
- The tubes became cloudy after vortexing:
- R13
- R5
- R14

- Incubated tubes at room temp for 3 minutes
- Centrifuged tubes for 15 minutes at 12,000rcf at room temp
- Prepared DNase treatment on ice:
- 24ul 10X reaction buffer
- 30ul DNAse
- 186ul RNase-free water
- After centrifugation tubes had phase separated:
- R13
- R5
- R14
- Was able to remove ~400ul of the top clearish layer into new 1.5mL tubes (tough to remove without getting other layers)
- Added 400ul 70% ethanol to each new tube and vortexed (note, no precipitates formed)
- Added 600ul to spin columns for each sample
- Centrifuged 15 seconds at 12,000 rcf (note, our centrifuges do not go 15 sec, set to 30 then stopped after 15 sec)
- Discarded flow through in trizol waste
- Added the remaining sample to their spin columns
- Centrifuged 15 seconds at 12,000 rcf
- Discarded flow through in trizol waste
- Added 350ul of wash buffer 1
- Centrifuged 15 seconds at 12,000 rcf
- Discarded flow through in trizol waste and changed collection tubes
- Added 80ul DNase mixture to each spin column and incubated at room temp for 12 minutes
- Added 60ml of 100% ethanol to wash buffer 2
- After incubation, added 350ul wash buffer 1
- Centrifuged 15 seconds at 12,000 rcf
- Discarded flow through in trizol waste and changed collection tubes
- Added 500ul wash buffer 2
- Centrifuged 15 seconds at 12,000 rcf
- Discarded flow through in trizol waste
- Again added 500ul wash buffer 2
- Centrifuged 15 seconds at 12,000 rcf
- Discarded flow through in trizol waste
- Centrifuged columns dry for 1 minute at 12,000rcf
- Transferred columns to recovery 1.5mL tubes
- Added 100ul RNase-free water to each column
- Incubated at room temp ~1minute
- Centrifuged for 2 minutes at 16,000rcf
- Aliquoited 5ul for QC and froze the big tubes in the -80
Qubit
- Followed qubit protocol
| Standard 1 | Standard 2 | Sample | RNA :( |
|---|---|---|---|
| 393 RFU | 8799 RFU | R13 | too low |
| - | - | R5 | too low |
| - | - | R14 | too low |
- Did not do a tapestation
Written on November 16, 2020