Simulating Double Digest to get Fragment Distributions
Using ddRADSeqTools, the Porites lutea genome, and the Pocillopora damicornis genome to simulate PstI and MspI restriction enzyme fragment distributions
Downloaded ddRADSeqTools from Github by clicking Clone or Download, and select Download Zip
This Downloaded the repo to my computer, which is probably not the best way to do this, but it worked. I then copied it to my KITT account.
# in KITT made a directory to work in
mkdir sim-rad-frags
Have to use my terminal to copy things to KITT
scp -r -P zzzz /Users/maggieschedl/Desktop/ddRADseqTools-master.zip mschedl@kitt.uri.edu:/home/mschedl/sim-rad-frags
Then I unzipped it and made the scrips exicutable by using the commands they have in the manual
unzip ddRADseqTools-master.zip
cd ddRADseqTools-master/Package
chmod u+x *.py
conda install NumPy #also the manual says you need this program installed
Then download the two genomes
Porites lutea
Pocillopora damicornis
scp -r -P zzzz /Users/maggieschedl/Desktop/porites_plut_final_2.1.fasta.gz mschedl@kitt.uri.edu:/home/mschedl/
scp -r -P zzzz /Users/maggieschedl/Desktop/poc_GCF_003704095.1_ASM370409v1_genomic.fna.gz mschedl@kitt.uri.edu:/home/mschedl/
Move them to storage then link them to the directory where the scripts are
mv porites_plut_final_2.1.fasta.gz /RAID_STORAGE2/mschedl/geenomes/
mv poc_GCF_003704095.1_ASM370409v1_genomic.fna.gz /RAID_STORAGE2/mschedl/geenomes/
cd sim-rad-frags/ddRADSeqTools-master/Package
ln -s /RAID_STORAGE2/mschedl/geenomes/* .
The program uses a text file with all the restriction enzymes and their cut sites as a reference, I went through the file and saw that they didn’t have MspI so I added it in
nano restrictionsites.txt
MspI;C*CGG # add this in alphabetical order
Then the rsitesearch.py script uses the rsitesearch-config.txt to get all the information on what genome and enzymes to use. This is the only script I used, because it “extracts the fragments resulting from an in silico digestion of a reference genome with two particular restriction endonucleases.”
I edited the config file to do Pocillopora first.
nano rsitesearch-config.txt
genfile=./poc_GCF_003704095.1_ASM370409v1_genomic.fna.gz # add in the pocillopora genome, the . because I linked it to this directory
fragsfile=./results/fragments.fasta # path of the fragments file
rsfile=./restrictionsites.txt # path of the restriction sites file
enzyme1=PstI # id of 1st restriction enzyme used in rsfile or its restriction site sequence
enzyme2=MspI # id of 2nd restriction enzyme used in rsfile or its restriction site sequence
minfragsize=150 # lower boundary of loci fragment's size
maxfragsize=700 # upper boundary of loci fragment's size
fragstfile=./results/fragments-stats.txt # path of the output statistics file
fragstinterval=25 # interval length of fragment size
plot=NO # statistical graphs: YES or NO
verbose=YES # additional job status info during the run: YES or NO
trace=NO # additional info useful to the developer team: YES or NO
Then you have to make the results directory for some reason before it runs
mkdir results
Then run the script
python rsitesearch.py
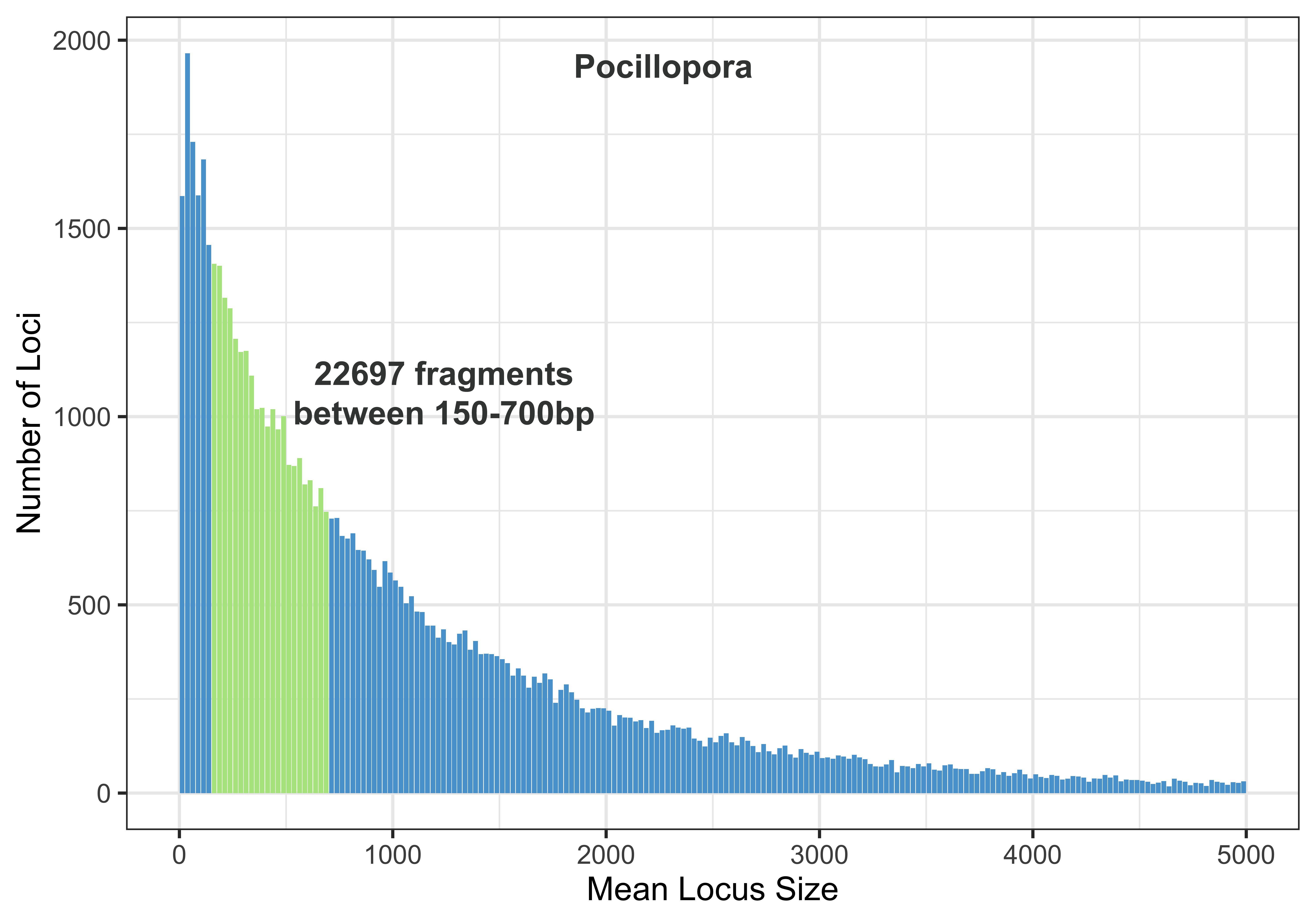
Output: 22679 fragments written
This means that there are 22679 fragments digested by both PstI and MspI in this Pocillopora genome that are between 150 and 700bp long.
Change the name of the directory because doing two of these
mv results results-poc
The names of the files it gives you are
fragments.fasta fragments-GC-distribution.csv poc-fragments-stats.csv poc-fragments-stats.txt
Copy the results to my computer to run them on R. I had to change the CSV file format in excel to make the figure in R
scp -r -P zzzz mschedl@kitt.uri.edu:/home/mschedl/sim-rad-frags/ddRADseqTools-master/Package/results/* /Users/maggieschedl/Desktop/URI/Maggie/PutnamPuritzMoorea
The CSV file doesn’t go into excel well, the columns are separated by ; instead of commas. I changed the fragments-stats.csv to a .txt by renaming it (it gives an error but do it anyways), then in excel in the Data tab, import from text and use that file. Follow the GUI and say that the fields are delimitated, then specify ; as the symbol. After that it makes a column for each field. But, the fragment interval values 1-25, 26-50 etc are not continuous, so those don’t work for the graph I wanted. So I created a new column to the right of the interval column to split it. Select the interval column and click text to column in the data tab. Again say delaminated and this time by - this should make two columns and I named them fragment_interval_start and fragment_interval_end. This way I could make another column mean_locus_size that was the average of those two columns. There’s probably a way to do this in R but 🤷🏻
Then I did the same thing for the Porites
nano rsitesearch-config.txt
genfile=./porites_plut_final_2.1.fasta.gz # file of the reference genome in fasta format
fragsfile=./results/fragments.fasta # path of the fragments file
rsfile=./restrictionsites.txt # path of the restriction sites file
enzyme1=PstI # id of 1st restriction enzyme used in rsfile or its restriction site sequence
enzyme2=MspI # id of 2nd restriction enzyme used in rsfile or its restriction site sequence
minfragsize=150 # lower boundary of loci fragment's size
maxfragsize=700 # upper boundary of loci fragment's size
fragstfile=./results/por-fragments-stats.txt # path of the output statistics file
fragstinterval=25 # interval length of fragment size
plot=NO # statistical graphs: YES or NO
verbose=YES # additional job status info during the run: YES or NO
trace=NO # additional info useful to the developer team: YES or NO
Saved and ran the script
mkdir results
python rsitesearch.py
mv results results-por
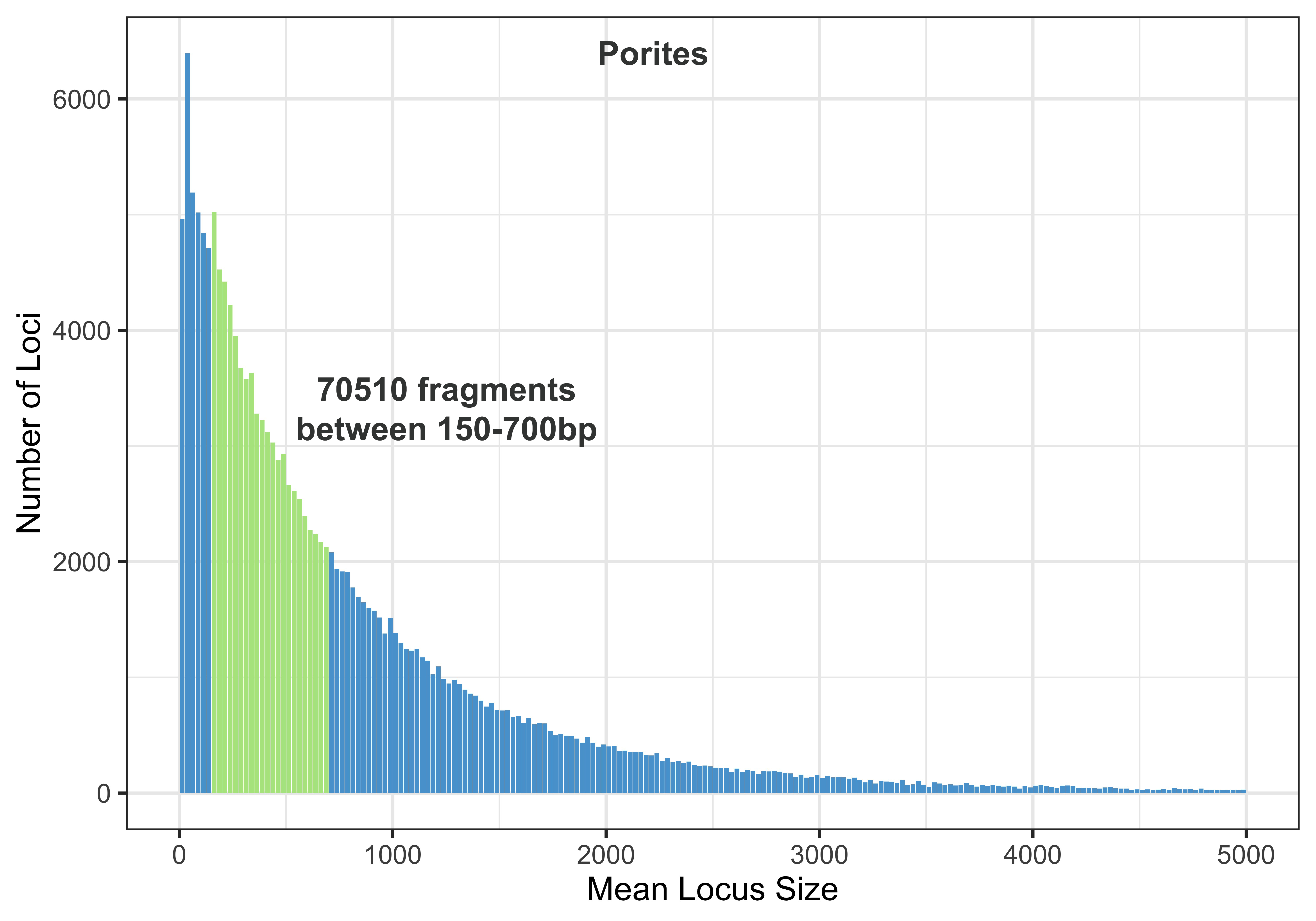
Output: 70510 fragments written
This means that there are 70510 fragments digested by both PstI and MspI in this Porites genome that are between 150 and 700bp long.
Then copied to my computer
scp -r -P zzzz mschedl@kitt.uri.edu:/home/mschedl/sim-rad-frags/ddRADseqTools-master/Package/results-por/* /Users/maggieschedl/Desktop/URI/Maggie/PutnamPuritzMoorea
Changed the csv file in the same way and renamed it for Porites
Using R to Make Figures of the Fragment Distributions
Load Packages
getwd()
library(ggplot2)
library(tidyverse)
Read the edited CSV files from ddRADSeqTools output
pocillopora_RAD_stats <- read.csv("POC-PstI-MspI-RAD-frags.csv")
porites_RAD_stat <- read.csv("POR-PstI-MspI-frags.csv")
Pocilliopora Fragments
Using mutate highlights the region between 150-700bp, then I also set the cut off of the x axis to be 5000, we aren’t looking that high anyways.
pocillopora_RAD_stats %>%
mutate(highlight_flag = ifelse(mean_loci_size > 150 & mean_loci_size < 700, T, F)) %>%
ggplot(aes(x=mean_loci_size, y=num_frags))+
geom_bar(aes(fill= highlight_flag), stat="identity") +
theme_bw(base_size=18) + scale_fill_manual(values = c('#58a2d3', '#b2e58e')) + theme(legend.position = "none") + xlab("Mean Locus Size") + ylab("Number of Loci") + xlim(0, 5000)
ggsave("pocillopora150-700frag.jpg", height=7, width=10, units="in", dpi=600)
Porites Fragments
porites_RAD_stat %>%
mutate(highlight_flag = ifelse(mean_loci_size > 150 & mean_loci_size < 700, T, F)) %>%
ggplot(aes(x=mean_loci_size, y=num_frags))+
geom_bar(aes(fill= highlight_flag), stat="identity") +
theme_bw(base_size=18) + xlim(0, 5000) + scale_fill_manual(values = c('#58a2d3', '#b2e58e')) + theme(legend.position = "none") + xlab("Mean Locus Size") + ylab("Number of Loci")
ggsave("porites150-700frag.jpg", height=7, width=10, units="in", dpi=600)