Invitrogen MethylMiner Methylated DNA Enrichment Kit Test
Testing MethylMiner Kit on Two Samples to Prepare for Pico-Methyl Seq
Using samples 13 and 14 from the shear test in the previous post (Acropora 221 and Montipora 1431) which were sheared to a peak around 500bp and have about 1µg of DNA
Using the Invitrogen Methylminer Methylated DNA Enrichment Kit with specifications for 1µg of input DNA and the SINGLE elution.
First step was to write out the steps for the day so I would know how much 1X Bind/Wash Buffer to dilute, as it comes in 5X in the kit, that way it was ready when it was needed (1600µl needed, 1700µl was made to account for any error)
Preparing Beads
- Pipetted up and down the Dynabeads M-280 Streptavidin to resuspend them
- Made 2 1.5mL tubes, each with 10µl of Dynabeads (the amount recommended for less than or equal to 1µg of DNA input)
- Brought volume up to a total of 100µl with 90µl of 1X bind/wash buffer to each tube
- Pipetted to mix
- Placed tubes on long magnet rack and removed and discarded supernatant when clear
- Removed tubes from rack and resuspended beads in 100µl 1X bind/wash buffer
- Placed tubes on long magnet rack and removed and discarded supernatant when clear
- Removed tubes from rack and resuspended beads in 100µl 1X bind/wash buffer
Coupling MBD-Biotin Protein to the Beads
- Thawed MBD-Biotin protein from -80 on ice
- Made 2 new 1.5mL tubes, each with 7µl of MBD-Biotin protein (amount recommended for 1µg of DNA input)
- Added 93µl of 1X bind/wash buffer to each tube to get up to a total of 100µl
- Transferred diluted protein to the washed bead tubes for a total volume of 200µl in each of 2 tubes
- Put samples on the rotisserie mixer for 1 hour at room temp
Washing MBD-Biotin-Coupled Beads
- Spun down briefly tubes from above
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Repeated: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Repeated again: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Finally: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
Capture Reaction
- To 2 new 1.5mL tubes, added 20µl each of 5X bind/wash buffer
- To the appropriate tube, added the 49µl of the DNA sample (221 and 1431 sheared)
- Brought volume up to a total of 100µl in each tube by adding 31µl of ultra pure water
- Transferred all of each diluted DNA sample to separate tubes with the MBD-Biotin bound beads for a total of 200µl in each tube and pipetted to mix
- Mixed on rotisserie mixer overnight at 4 degrees C in the cold room
Aug 2nd 2019, Removing Non-Captured DNA
- Again started by writing out protocol to calculate how much 1X bind/wash buffer to dilute (800µl needed)
- Took tubes out of the cold room rotisserie and spun down briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in tubes labeled non-captured DNA and sample number
- Resuspended beads in 200µl of 1X bind/wash buffer and placed on rotisserie mixer for 3 minutes
- Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in tubes labeled wash and the sample number
- Repeated: added 200µl 1X bind/wash buffer and placed on rotisserie mixer for 3 minutes
- Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in the same tubes labeled wash and the sample number for a total of 400µl in each tube
Single Fraction Elution
- Resuspended beads in 200µl High Salt Elution buffer
- Incubated on rotisserie mixer for 3 minutes
- Spun down tubes breifly and placed on magnet rack for 1 minute
- Removed supernatant when clear and SAVED in a new 1.5mL tube labeled captured DNA and the sample number
- Repeated: added 200µl High Salt Elution Buffer to resuspend beads in each tube
- Incubated tubes on rotisserie mixer for 3 minutes
- Spun down tubes briefly and placed on magnet rack for 1 minute
- Removed clear supernatant and SAVED in each sample tube for captured DNA, for a total of 400µl in each tube
Ethanol Precipitation
Each tube for ethanol precipitation gets 1µl of glycogen (co-precipitator), 1/10th the volume of the sample of 3M sodium acetate pH 5.2, and 2 volumes of the sample 100% ethanol, so…
| Sample | vol glycogen (µl) | vol sodium acetate (µl) | vol 100% EtOH (µl) |
|---|---|---|---|
| 221 Non-Captured (NC) | 1 | 20 | 400 |
| 1431 Non-Captured (NC) | 1 | 20 | 400 |
| 221 Washed Non-Captured (W) | 1 | 40 | 800 |
| 1431 Washed Non-Captured (W) | 1 | 40 | 800 |
| 221 Captured (C) | 1 | 40 | 800 |
| 1431 Captured (C) | 1 | 40 | 800 |
Note, I did not do this in the hood but I should have, the 3M Sodium Acetate pH 5.2 should be kept in the acid cabinet and used in the hood
- Vortexed to mix and spun down
- Placed tubes in the -80 freezer for 2 hours
- Set centrifuge to 4 degrees C (ours can actually get to 1 degree so next time do that) with about 15 min time left and let it run to get down to temp
- Prepared 4mL of 70% EtOH and put in -20 to chill down
- Took sample tubes out of -80 and centrifuged for 15 minutes at 4 degrees C and 14,000 rcf
- Took tubes out of centrifuge one at a time to keep cold, looked at tubes to see if there was a pellet. The non-captured 1431 sample did not have a visible pellet. In all these steps tubes were taken out of the centrifuge one at a time to keep them cold
- Removed the supernatant and discarded very carefully, and I pretended like sample 1431 NC had a pellet and left a small amount of supernatant in the tube
- Added 500µl of cold 70% EtOH to each tube carefully
- Centrifuged for 5 minutes at 4 degrees C and 14,000 rcf
- Very carefully removed supernatant from pellet, once again pretending like 1431 NC had a pellet
- Centrifuged tubes for 5 minutes at 4 degrees C at 14,000 rcf
- Removed any remaining supernatant as best as possible with the smallest pipette tip and making sure to not removed the pellet
- Air dried pellet for 3 minutes
- Resuspended pellets in 25µl of ultra pure water
High Sensitivity DNA Qubit
- Followed Qubit protocol for HS reagents
| Sample | Standard 1 (RFU) | Standard 2 (RFU) | 1st reading (ng/ul) | Second reading (ng/ul) | Average ng/ul |
|---|---|---|---|---|---|
| 221 NC | 66.3 | 23643 | 18 | 18 | 18 |
| 1431 NC | 66.3 | 23643 | 40 | 40.4 | 40.2 |
| 221 W | 66.3 | 23643 | .5 | .503 | .501 |
| 1431 W | 66.3 | 23643 | .63 | .632 | .631 |
| 221 C | 66.3 | 23643 | 3.28 | 3.3 | 3.29 |
| 1431 C | 66.3 | 23643 | 2.24 | 2.24 | 2.24 |
- D5000 TapeStation following protocol
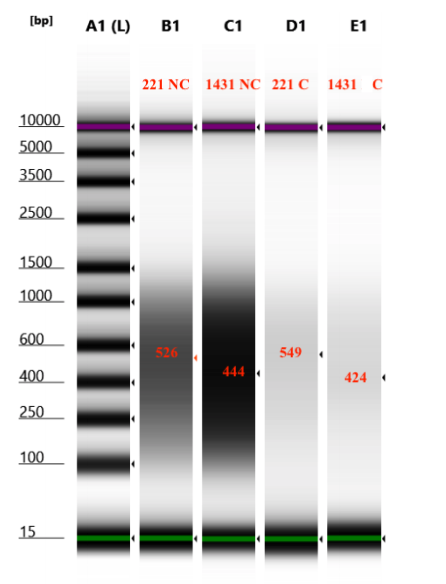
Looks like there isn’t a significant difference in the size captured or not captured by the beads: good!