Reconditioning PCR of KBay WGBS Libraries
Doing a 1 Cycle Reconditioning PCR on KBay WGBS Libraries That Looked Over-Amplified/Overly Broad Peaks
Goal Try 1 cycle PCR on some libraries with a broad or double peak to remove potential PCR bubbles, as stated here
Results All libraries have normal peaks after PCR
Takeaways Looks like this process works really well, all peaks look pretty much normal now. However, this process does use up reagents from the library prep kit that are limiting to us
10 Samples were chosen because their tapestations showed either double peaks or very broad peaks
For each sample I repeated the 2nd amplification from the WGBS protocol. The samples had more than 12ul volume in the final libraries, so if this did not work I knew I had some saved that we could use. The PCR program was copied and the program changed to have only 1 cycle. This is called 1 PICO METHYL AMP 2 in the thermocycler program directory.
| Extraction-ID | Library Volume | KAPA Mix Volume | Primer Pair | Volume Primer |
|---|---|---|---|---|
| 22 | 12ul | 13ul | 2 | 1ul |
| 26 | 12ul | 13ul | 3 | 1ul |
| 38 | 12ul | 13ul | 11 | 1ul |
| 34 | 12ul | 13ul | 18 | 1ul |
| 50 | 12ul | 13ul | 21 | 1ul |
| 33 | 12ul | 13ul | 24 | 1ul |
| 54 | 12ul | 13ul | 26 | 1ul |
| 39 | 12ul | 13ul | 27 | 1ul |
| 56 | 12ul | 13ul | 31 | 1ul |
| 59 | 12ul | 13ul | 40 | 1ul |
After the PCR (13 minutes), I performed the normal 1X bead cleanup as outlined in the protocol. The libraries were resuspended in 17ul elution buffer.
Broad Range dsDNA Qubit
- Followed Qubit protocol
| Sample | Reading 1 (ng/ul) | Reading 2(ng/ul) | Average (ng/ul) |
|---|---|---|---|
| S1 | 178 RFU | - | - |
| S2 | 20137 RFU | - | - |
| 22 | 54.4 | 54.2 | 54.3 |
| 26 | 53.2 | 53 | 53.1 |
| 38 | 54.6 | 54.8 | 54.7 |
| 34 | 53.4 | 53 | 53.2 |
| 50 | 46 | 45.6 | 45.8 |
| 33 | 50.4 | 50.2 | 50.3 |
| 54 | 47.8 | 48.2 | 48 |
| 39 | 51 | 50.8 | 50.9 |
| 56 | 37.8 | 37.6 | 37.5 |
| 59 | 51.6 | 51 | 51.3 |
D5000 TapeStation
- Followed TapeStation protocol
- I used two tapes
- Link 1
- Link 2
Example before and after traces, clearly two peaks visible in the first one:
Before

After
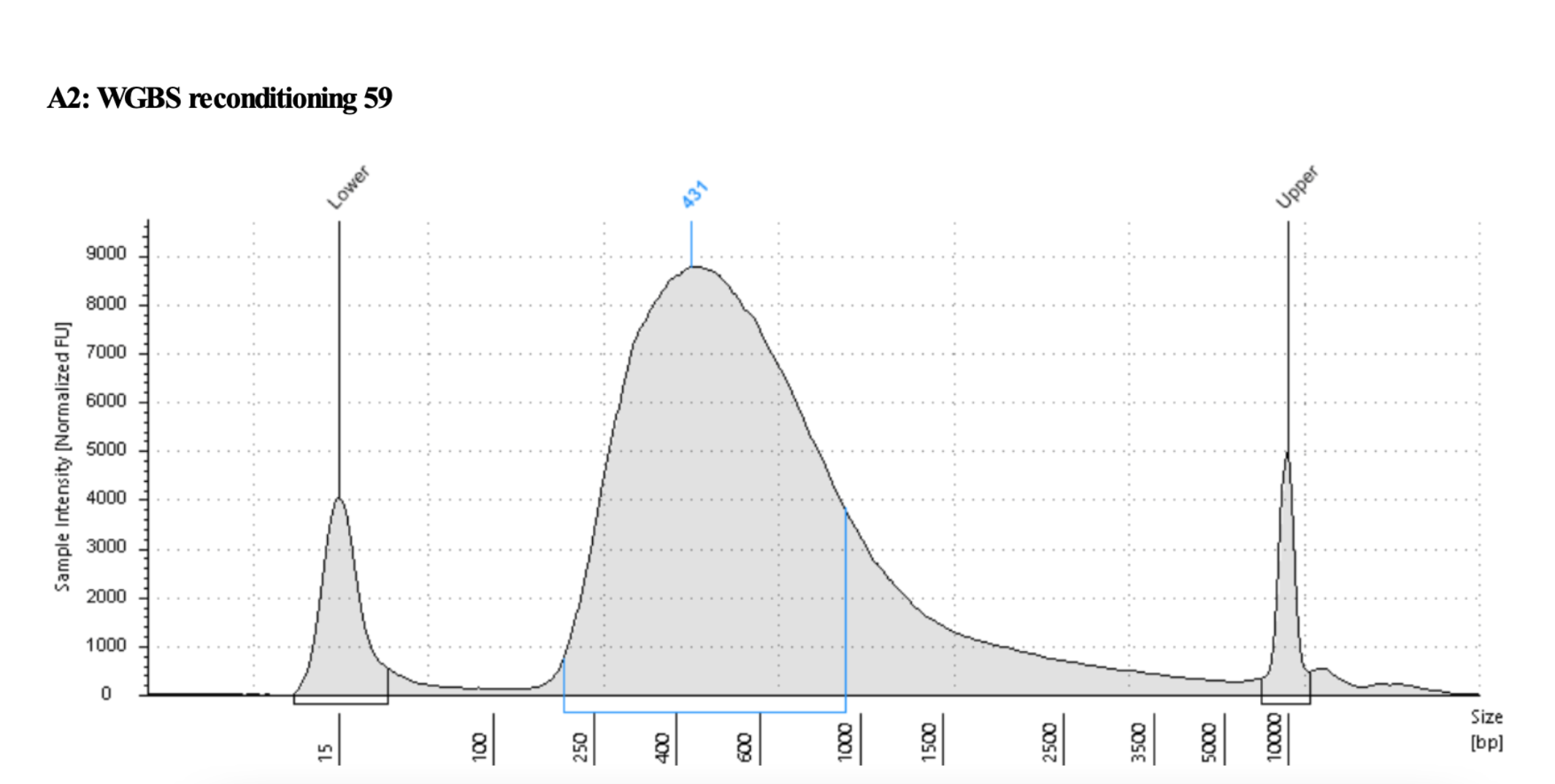
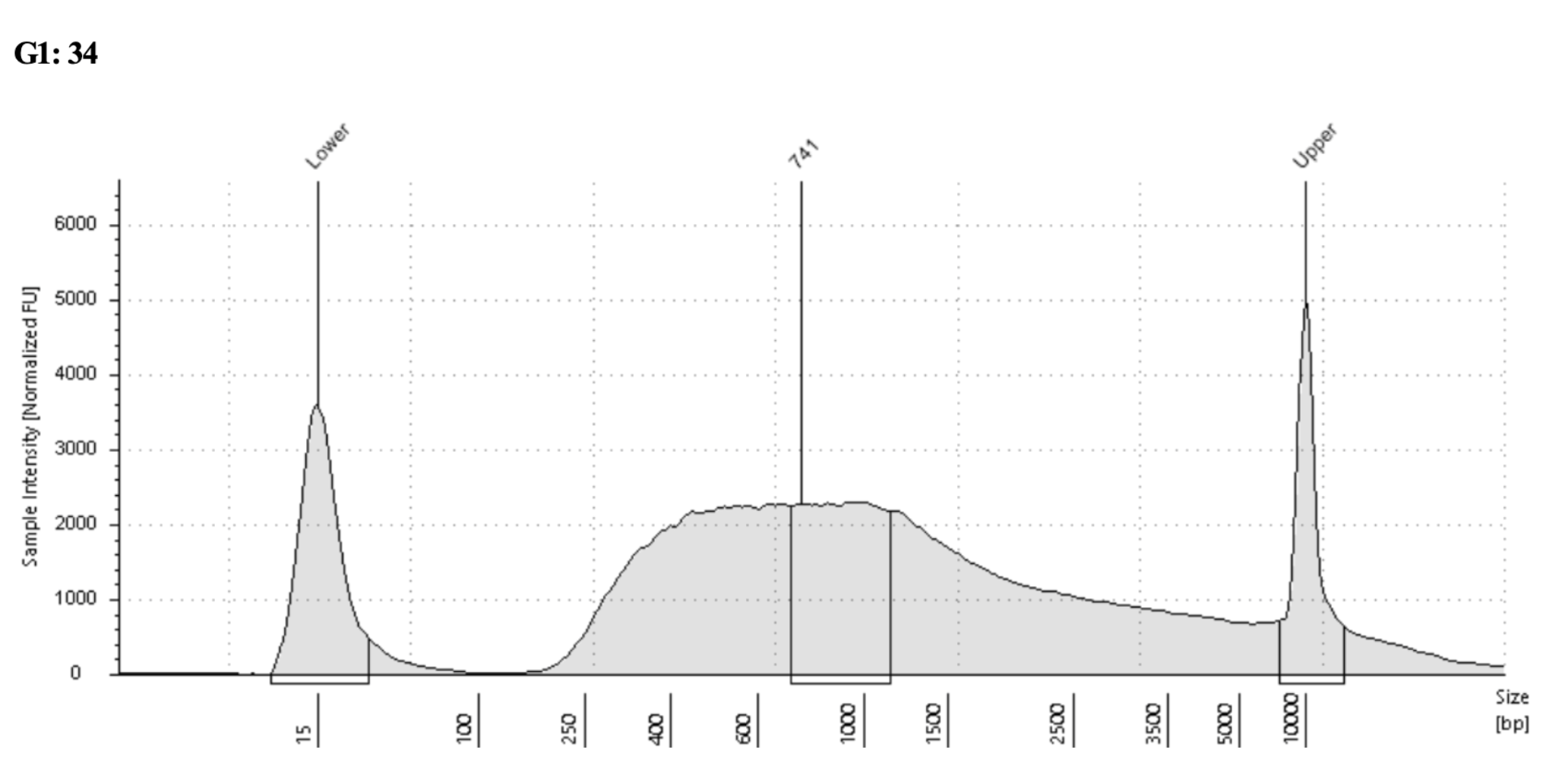
Example before and after traces, sample that may have two peaks or just one big one:
Before
After
