Using the Zymo Pico Methyl Seq Library Prep Kit for the 1st 16 of Danielle’s Pocillopora DNA Samples for Whole Genome Bisulfite Sequencing
Goal Library prep 16 samples for WGBS
Results 15 samples were prepped successfully
Takeaways The sample that didn’t get prepped well was in a spin column that looked strange after the first spin. That may have caused a problem/the failure. Also it is super helpful to do the bisulfite conversion the day before to save ~2 hours. Even so doing 16 samples from BS cleanup takes an entire day of lab work and is teadious and long.
This library prep followed the exact protocol for the Zymo Pico Methyl Seq Kit from me. See that protocol for detailed descriptions of each steps. Tables and values specific for this prep are included below.
2020 09 24 DNA Dilution to 1ng/ul of all samples
| Coral ID |
DNA for dilution to 1ng/ul (ul) |
10mM Tris HCl for dilution to 1ng/ul (ul) |
| E3 |
2 |
19.7 |
| E7 |
2 |
35.8 |
| E14 |
2 |
12.86 |
| C26 |
2 |
12.96 |
| C31 |
2 |
28.9 |
| C29 |
2 |
7.62 |
| E13 |
2 |
29.7 |
| C20 |
2 |
39.8 |
| C18 |
2 |
27.4 |
| C24 |
2 |
23.4 |
| E10 |
2 |
20.7 |
| C27 |
2 |
5.02 |
| C22 |
2 |
13.54 |
| E11 |
2 |
11.64 |
| E4 |
2 |
8.16 |
| C28 |
2 |
42.4 |
| E6 |
2 |
14.66 |
| C21 |
2 |
3.3 |
| E8 |
2 |
3.98 |
| E12 |
2 |
33.1 |
| E5 |
2 |
33.4 |
| C30 |
2 |
8.62 |
| E1 |
2 |
13.1 |
| C23 |
2 |
19.8 |
| E16 |
2 |
22.8 |
| C19 |
2 |
16.36 |
| C25 |
2 |
37.4 |
| E2 |
2 |
20.6 |
| E9 |
2 |
20.8 |
| C32 |
2 |
5.08 |
| E15 |
2 |
5.42 |
| C17 |
2 |
35.4 |
2020 09 24 Bisulfite Conversion
- Samples used for first prep:
- E3
- E7
- E14
- C26
- C31
- C29
- E13
- C20
- C18
- C24
- E10
- C27
- C22
- E11
- E4
- C28
- Followed exact steps as in protocol
- Once the thermocycler program was done the samples were put in the 4 degree fridge overnight
2020 09 25
Post-BS Conversion cleanup
- Followed exact steps as in the protocol
- note: I noticed the spin column for E7 had residue all up one side of it after the first spin. I don’t know if it had been there before spinning and I had just not noticed. It sort of looked like the filter substance had gotten up the side of the spin column
Amplification with Prep-Amp Primers
- Followed exact steps as in the protocol
- Priming Master Mix calculations (PMM):
- 2ul PrepAmp Buffer * 16.5 = 33ul
- 1ul PrepAmp Primer * 16.5 = 16.5ul
- PrepAmp Master Mix calculations (PAMM):
- 1ul PrepAmp Buffer * 16.5 = 16.5ul
- 3.75ul PrepAmp PreMix * 16.5 = 61.875ul
- 0.3ul PrepAmp Polymerase * 16.5 = 4.95ul
- Dilution calculation of PrepAmp Polymerase to add 0.5ul:
- 0.3 PrepAmp Polymerase * 16.5 = 4.95ul
- 0.2ul DNA elution buffer * 16.5 = 3.3ul
DNA Clean and Concentrator
- Followed exact steps as in the protocol
First Amplification
- Followed exact steps as in the protocol
- 1st Amp Master Mix calculation:
- 12.5ul Library Amp Mix * 16.5 = 206.25ul
- 1ul Library Amp Primers * 16.5 = 16.5ul
Second DNA Clean and Concentrator
- Followed exact steps as in the protocol
Second Amplification with Index Primers
- Followed exact steps as in the protocol
- Thermocycler program had 12 cycles
- Table for components in tubes for amplifications:
| Sample |
Volume DNA (ul) |
Volume Library Amp Mix (ul) |
Volume i5 Primer (10uM) |
Volume i7 Primer (10uM) |
| E3 |
12 |
14 |
1ul i5_ZM_UDI001 |
1ul i7_ZM_UDI001 |
| E7 |
12 |
14 |
1ul i5_ZM_UDI002 |
1ul i7_ZM_UDI002 |
| E14 |
12 |
14 |
1ul i5_ZM_UDI003 |
1ul i7_ZM_UDI003 |
| C26 |
12 |
14 |
1ul i5_ZM_UDI004 |
1ul i7_ZM_UDI004 |
| C31 |
12 |
14 |
1ul i5_ZM_UDI005 |
1ul i7_ZM_UDI005 |
| C29 |
12 |
14 |
1ul i5_ZM_UDI006 |
1ul i7_ZM_UDI006 |
| E13 |
12 |
14 |
1ul i5_ZM_UDI007 |
1ul i7_ZM_UDI007 |
| C20 |
12 |
14 |
1ul i5_ZM_UDI008 |
1ul i7_ZM_UDI008 |
| C18 |
12 |
14 |
1ul i5_ZM_UDI009 |
1ul i7_ZM_UDI009 |
| C24 |
12 |
14 |
1ul i5_ZM_UDI010 |
1ul i7_ZM_UDI010 |
| E10 |
12 |
14 |
1ul i5_ZM_UDI011 |
1ul i7_ZM_UDI011 |
| C27 |
12 |
14 |
1ul i5_ZM_UDI012 |
1ul i7_ZM_UDI012 |
| C22 |
12 |
14 |
1ul i5_ZM_UDI013 |
1ul i7_ZM_UDI013 |
| E11 |
12 |
14 |
1ul i5_ZM_UDI014 |
1ul i7_ZM_UDI014 |
| E4 |
12 |
14 |
1ul i5_ZM_UDI015 |
1ul i7_ZM_UDI015 |
| C28 |
12 |
14 |
1ul i5_ZM_UDI016 |
1ul i7_ZM_UDI016 |
1X Bead Clean
- Followed exact steps as in protocol
- Samples were Qubited immediately so they were put on an ice bucket not frozen yet
Broad Range dsDNA Qubit
| Sample |
Reading 1 (ng/ul) |
Reading 2(ng/ul) |
Average (ng/ul) |
| Standard 1 |
192 RFU |
- |
- |
| Standard 2 |
22769 RFU |
- |
- |
| E3 |
21 |
20.8 |
20.9 |
| E7 |
too low |
- |
- |
| E14 |
21.8 |
21.6 |
21.7 |
| C26 |
17.1 |
16.9 |
17 |
| C31 |
9.84 |
9.88 |
9.86 |
| C29 |
11.9 |
11.9 |
11.9 |
| E13 |
20.8 |
21 |
20.9 |
| C20 |
17.5 |
17.5 |
17.5 |
| C18 |
13.2 |
13.2 |
13.2 |
| C24 |
14.1 |
14 |
14.05 |
| E10 |
18.4 |
18.3 |
18.35 |
| C27 |
18.9 |
19 |
18.95 |
| C22 |
10.4 |
10.4 |
10.4 |
| E11 |
18.8 |
19 |
18.9 |
| E4 |
18 |
17.9 |
17.95 |
| C28 |
6.82 |
6.67 |
6.79 |
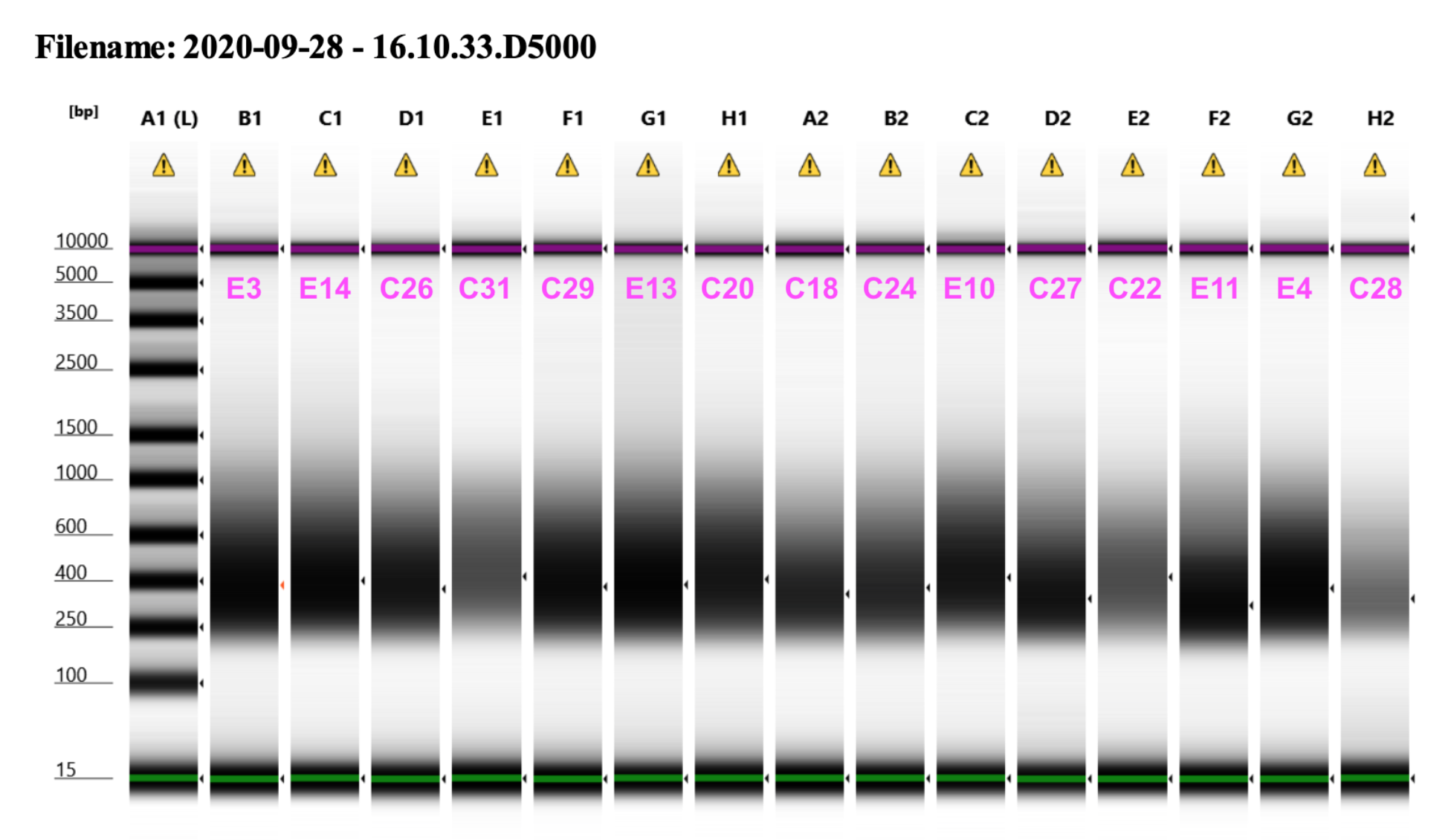
2020 09 28 D5000 TapeStation
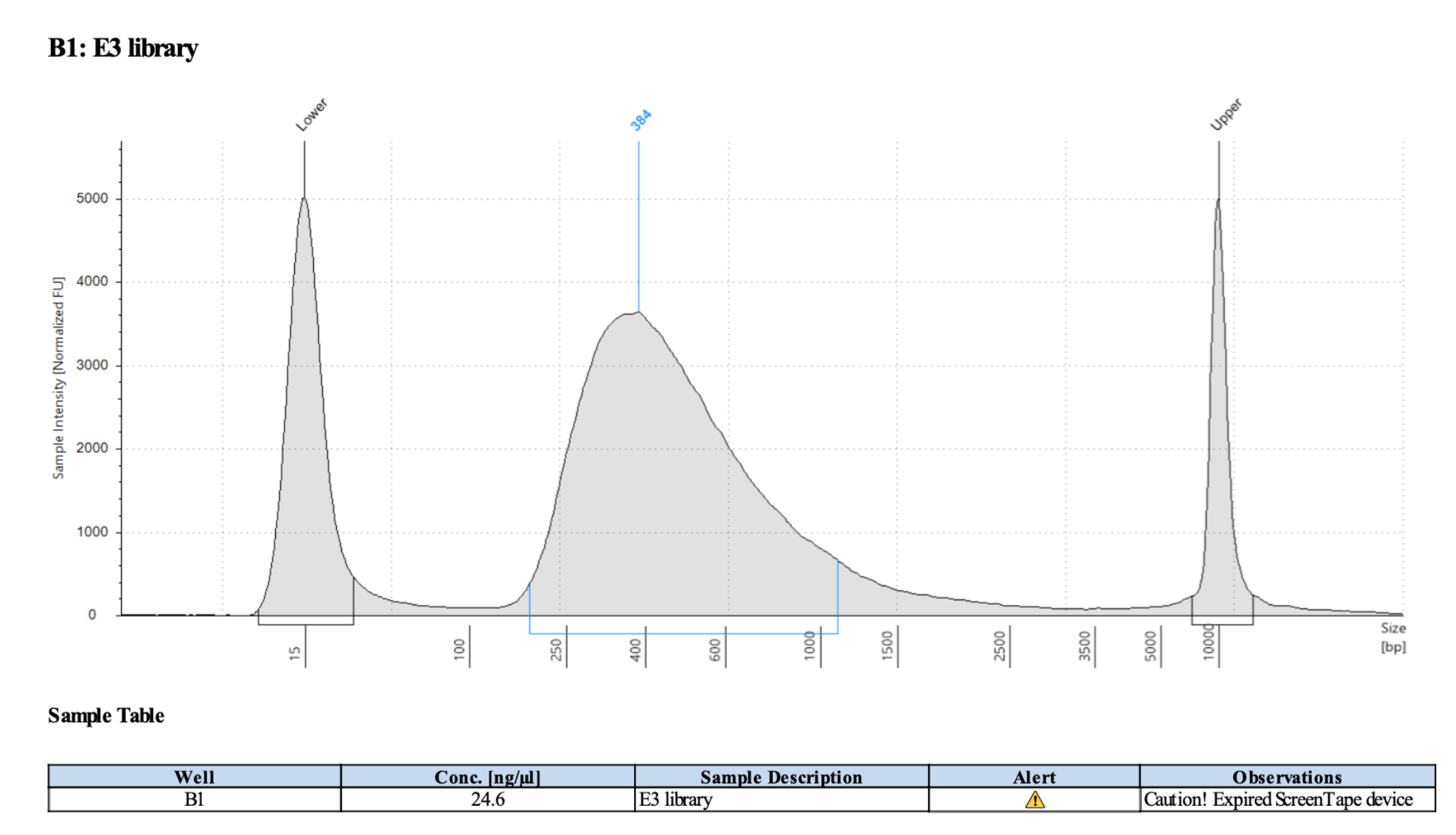
Representative library trace:
Samples and Index Sequences
| Coral ID |
i7 bases |
i5 bases |
| E3 |
CCGCGGTT |
AGCGCTAG |
| E7 |
TTATAACC |
GATATCGA |
| E14 |
GGACTTGG |
CGCAGACG |
| C26 |
AAGTCCAA |
TATGAGTA |
| C31 |
ATCCACTG |
AGGTGCGT |
| C29 |
GCTTGTCA |
GAACATAC |
| E13 |
CAAGCTAG |
ACATAGCG |
| C20 |
TGGATCGA |
GTGCGATA |
| C18 |
AGTTCAGG |
CCAACAGA |
| C24 |
GACCTGAA |
TTGGTGAG |
| E10 |
TCTCTACT |
CGCGGTTC |
| C27 |
CTCTCGTC |
TATAACCT |
| C22 |
CCAAGTCT |
AAGGATGA |
| E11 |
TTGGACTC |
GGAAGCAG |
| E4 |
GGCTTAAG |
TCGTGACC |
| C28 |
AATCCGGA |
CTACAGTT |
| E6 |
TAATACAG |
ATATTCAC |
| C21 |
CGGCGTGA |
GCGCCTGT |
| E8 |
ATGTAAGT |
ACTCTATG |
| E12 |
GCACGGAC |
GTCTCGCA |
| E5 |
GGTACCTT |
AAGACGTC |
| C30 |
AACGTTCC |
GGAGTACT |
| E1 |
GCAGAATT |
ACCGGCCA |
| C23 |
ATGAGGCC |
GTTAATTG |
| E16 |
ACTAAGAT |
AACCGCGG |
| C19 |
GTCGGAGC |
GGTTATAA |
| C25 |
CTTGGTAT |
CCAAGTCC |
| E2 |
TCCAACGC |
TTGGACTT |
| E9 |
CCGTGAAG |
CAGTGGAT |
| C32 |
TTACAGGA |
TGACAAGC |
| E15 |
GGCATTCT |
CTAGCTTG |
| C17 |
AATGCCTC |
TCGATCCA |