8 Mo'orea Coral Sample Extractions
8 DNA only extractions from Porites and Pocillopora Corals from Mo’orea
Using the Zymo Quick-DNA Miniprep Plus kit
Sample Prep
| Sample # | Site # | Date Collected | Type |
|---|---|---|---|
| 175 | 5 | 2018/03/13 | Pocillopora verrucosa |
| 177 | 5 | 2018/03/13 | Pocillopora verrucosa |
| 178 | 5 | 2018/03/13 | Pocillopora verrucosa |
| 185 | 5 | 2018/03/13 | Pocillopora verrucosa |
| 197 | 5 | 2018/03/13 | Massive Porites |
| 200 | 5 | 2018/03/13 | Massive Porites |
| 205 | 5 | 2018/03/13 | Massive Porites |
| 303 | 5 | 2018/03/13 | Massive Porites |
- New Beads were poured into sample tubes. The new beads should be easier to pipette the liquid out of them as the do not get sucked up by the p20
- Samples were homogenized by vortexing for ~30 seconds. Porites samples were homogenized for an extra ~30 seconds
- Most of the liquid from the tubes was removed by pipetting. This was about 450µl Note that sample 205 did not homogenize noticeably, and did not look like a Porites. The tubes contained a small amount of liquid and un-homogenized tissue left, so those were put back into the -20
- Following recommendations for samples in DNA/RNA Shield from the kit protocol, 225µl of Solid Tissue Buffer and 15µl of Proteinase K were added to each sample
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
DNA Extraction
- Centrifuged all tubes at 12,000 rcf for 1 minute to pellet any debris and beads
- Removed supernatant into new 1.5mL tubes
- Here the proper step is to add 1 volume G-DNA Binding Buffer, but 1 volume (~690µl) G-DNA Wash Buffer was added on accident, this mistake was not noticed until further steps down, but the entire extraction was still processed
- 700µl of sample was added to the kit spin column and centrifuged at 12,000 rcf for 1 minute
- Collection tubes were discarded
- The rest of the coral samples were run through the column in the same way
- Added 400µl DNA Pre-Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 700µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 200µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the collection tube
- Columns were transferred to 1.5mL tubes
- Added 50µl warmed 70 degrees C 10mM Tris-HCl directly to the column filter and incubated at room temp for 5 minutes
- Centrifuged for 1 minute at 12,000 rcf
- Repeated steps 11 and 12 one more time
Qubit proceeded immediately to check if the mistake resulted in a failed extraction.
Qubit
- Broad Range dsDNA Qubit protocol
- All samples were read twice
- Standards weren’t recorded in this Qubit because the recorder assumed no DNA would be present
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA |
|---|---|---|---|---|---|
| 175 | - | - | 4.04 | 3.96 | 4 |
| 177 | - | - | 5.36 | 5.3 | 5.33 |
| 178 | - | - | 28.8 | 28.6 | 28.7 |
| 185 | - | - | 6.58 | 6.54 | 6.57 |
| 197 | - | - | 44.0 | 44.6 | 44.3 |
| 200 | - | - | 54.6 | 54.4 | 54.5 |
| 205 | - | - | too low | too low | - |
| 303 | - | - | 71.6 | 71.6 | 71.6 |
There is DNA! But, some of the values are very low so I wanted to try extracting from what was left in the tubes with the beads. I also am not sure that the quality is good so I need to run a gel. Samples were stored in 4 degree until the gel could be run the next day.
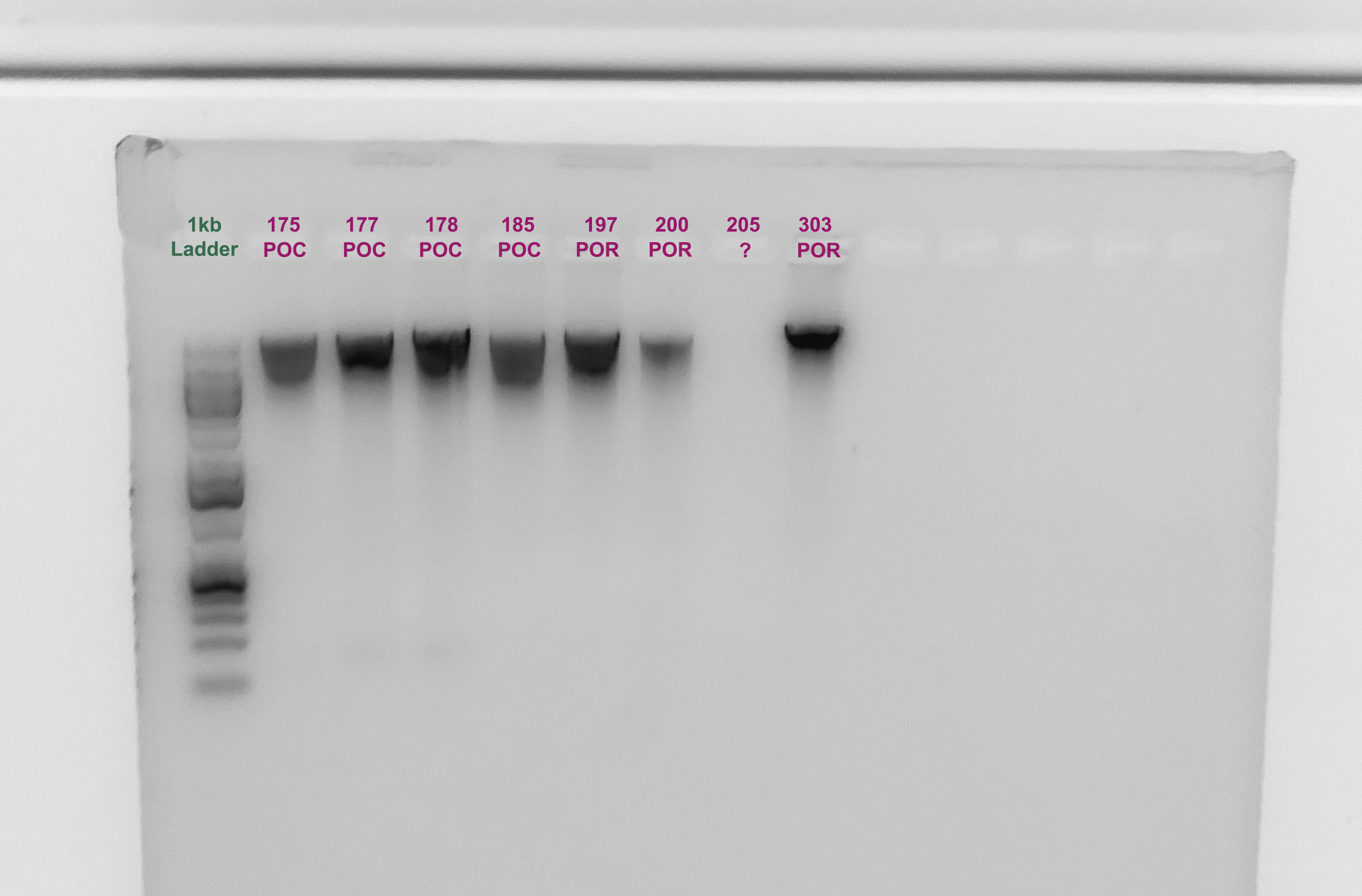
Gel
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
The quality of the DNA is good actually, but the quantify is low for some samples. Notably 7 didn’t show up, but that is concurrent with the Qubit.
04-11-19 Re-Extraction
- Added 250µl of DNA/RNA Shield to all of the tubes with the beads and remaining samples
- Samples were homogenized again for about a minute. Sample 205 still did not break up
- To make sure ALL the sample got out to maximize DNA yield, I used the sieve method again
- Sieves/strainers were placed in new labeled 1.5mL tubes
- About 1/3 of the sample was poured into the sieves and centrifuged briefly as the lid does not fit on the centrifuge
- The liquid sample went through into the tube but most of the beads stayed in the sieve
- Repeated pouring the rest of the sample into the sieve and centrifuging quickly
- 150µl of Solid Tissue Buffer and 10µl of Proteinase K were added to each sample
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
DNA Extraction
- Centrifuged all tubes at 12,000 rcf for 1 minute to pellet any debris and beads that got through the sieve
- Removed supernatant into new 1.5mL tubes
- Added 1 volume (460µl) Genomic Binding Buffer to each tube, vortexed and spun down
- 700µl, or all for the mussels, of sample was added to the kit spin column and centrifuged at 12,000 rcf for 1 minute
- Collection tubes were discarded
- The rest of the coral samples were run through the column in the same way
- Added 400µl DNA Pre-Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 700µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 200µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the collection tube
- Columns were transferred to 1.5mL tubes
- Added 50µl warmed 70 degrees C 10mM Tris-HCl directly to the column filter and incubated at room temp for 5 minutes
- Centrifuged for 1 minute at 12,000 rcf
- Repeated steps 11 and 12 one more time
- Sample tubes were labeled and stored in the 4 degree fridge to quantify the next day
Qubit
- Broad Range dsDNA Qubit protocol
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA |
|---|---|---|---|---|---|
| 175 | 187 | 20375 | 32 | 32.2 | 32.1 |
| 177 | 187 | 20375 | 36.4 | 36.6 | 36.5 |
| 178 | 187 | 20375 | 60.6 | 61.4 | 61 |
| 185 | 187 | 20375 | 38.4 | 38.4 | 38.4 |
| 197 | 187 | 20375 | 41.4 | 41.6 | 41.5 |
| 200 | 187 | 20375 | 15.3 | 15.5 | 15.4 |
| 205 | 187 | 20375 | too low | too low | - |
| 303 | 187 | 20375 | 49 | 49.8 | 49.4 |
Still no DNA for 205. When looking at the tissue sample, it does not look like a massive Porites at all, more like algae. It might be a miss-labeled tube.
All tubes from these extraction are labeled A for the first extraction and B for the second and stored in the -20.
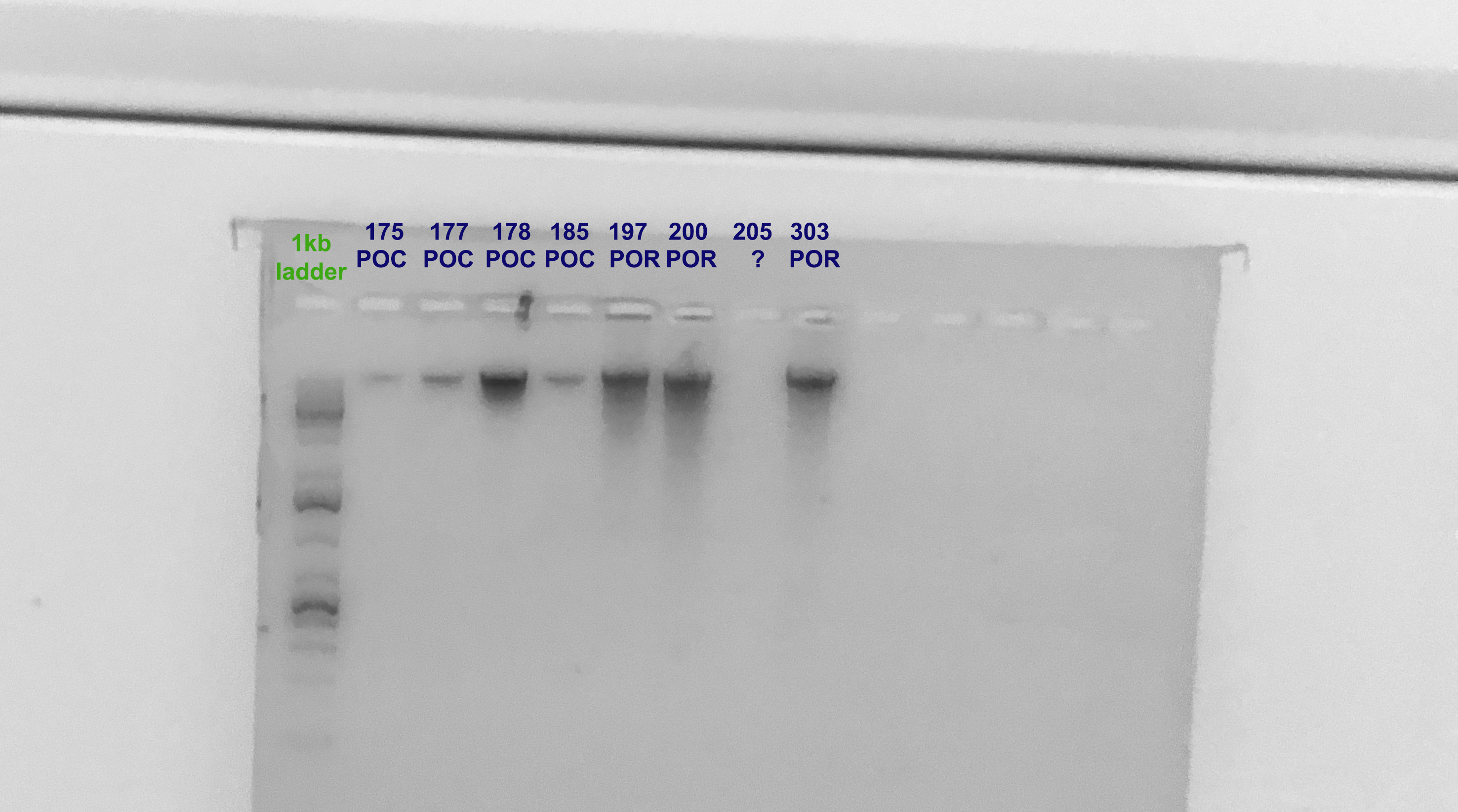
Gel Verification
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol