Clean-up, Quantification, and Prep for mtORF Test Sequence Samples
Bead Clean, Qubit Quantification, and Dilution and Aliquots for Sanger Sequencing of mtORF Amplifications
Notes
- Primer chosen for use during sequencing: RORF. Only one primer is used in Sanger Sequencing
- 3 samples from each project are being used for test sequencing. Samples E2, E6, and E10 from Danielle, and 65, 190, and 234 from the Mo’orea connect project. Samples E2, E6, E10, 65, and 190 were amplified here and 234 was amplified here
1X Bead Cleanup
- Only samples being sequenced were cleaned to save reagents
- The negative control reactions from here were also cleaned and quantified
- Steps were done at the post PCR bench
- Combined triplicate reactions from this amplification back together (~75ul)
- Used one 100ul reaction for 234
- Added 1X room temp resuspended KAPA Pure Beads to each well and pipetted to mix 10 times
- Placed tubes on shaker for 15 minutes at 200rpm
- Prepared fresh 80% ethanol
- Placed tubes on magnet rack after shaking and waited until liquid became clear
- Removed liquid from tubes without disturbing beads
- Added 200ul fresh 80% ethanol to each tube
- Removed liquid from tubes without disturbing beads
- Added 200ul fresh 80% ethanol to each tube
- Removed all liquid from tubes without disturbing beads
- Waited ~2 minutes for residual ethanol to evaporate but not for beads to dry
- Resuspended beads in 50ul ultra pure water
- Placed tubes on shaker 5 min 200 rpm
- After incubation placed tubes on magnet and waited until liquid became clear
- Removed liquid from tubes without disturbing beads and placed into new labeled 8-strip tubes
Broad Range Qubit
- Followed qubit protocol
- Included the control so 7 samples, 2 standards, and .5 for error
- Control was included because there seemed like there could be some contamination amplification in it
| Sample | Reading 1 | Reading 2 | Average DNA (ng/ul) |
|---|---|---|---|
| standard 1 | 175 | - | - |
| standard 2 | 18311 | - | - |
| E2 | 44 | 43.8 | 43.9 |
| E6 | 42.4 | 42.4 | 42.4 |
| E10 | 31.6 | 31.4 | 31.5 |
| 65 | 61.4 | 41.2 | 61.3 |
| 190 | 60.2 | 59.4 | 59.8 |
| 234 | 95.4 | 95.6 | 95.5 |
| Control | too low | - | - |
Dilution and Prep for Sequencing
- For sequencing I need 12ul (6 * 2) of 3.2uM RORF primer
- I also need 1.25ng for ever 100bp to be sequenced * 2 of each sample
- Things are multiplied by two for duplicate sequencing reactions
- The mtORF product should be ~1000 bp, so 1000 / 100 * 1.25 * 2 = 25ng of each sample
- 25ng is still less than 1ul for every sample, so I did a 1:10 dilution for each one
- Added 18ul of ultra pure water to 6 new strip tubes, and added 2ul of each cleaned DNA to their corresponding well
- Pipetted 25ng of each sample from diluted stock to new strip tubes
- Increased volume in tubes to 10ul with ultra pure water
| Sample | GSC sample code | Dilution conc. | ng needed | Volume diluted PCR product | volume ultra pure H2O |
|---|---|---|---|---|---|
| E2 | MHP01 | 4.4 | 25 | 5.68 | 4.32 |
| E6 | MHP02 | 4.2 | 25 | 5.95 | 4.05 |
| E10 | MHP03 | 3.1 | 25 | 8.06 | 1.94 |
| 65 | MHP04 | 6.1 | 25 | 4.1 | 5.9 |
| 190 | MHP05 | 6 | 25 | 4.16 | 5.84 |
| 234 | MHP06 | 9.5 | 25 | 2.63 | 7.37 |
- Made 14ul of diluted 10uM stock RORF to 3.2uM
- Added 9.52ul of ultra pure water to 1 PCR tube
- Added 4.48ul of 10uM RORF primer to tube
- Vortex and spin down to mix
- Added 2ul of 3.2uM primer to each of the 6 strip tubes diluted for sequencing
- Stored prepared samples in -20 freezer
Gel of 1:10 Diluted Cleaned PCR Products
- Asked Kevin Wong to add the diluted samples to a gel he was running to see if the previous gels looked strange because there was too much DNA
- Post
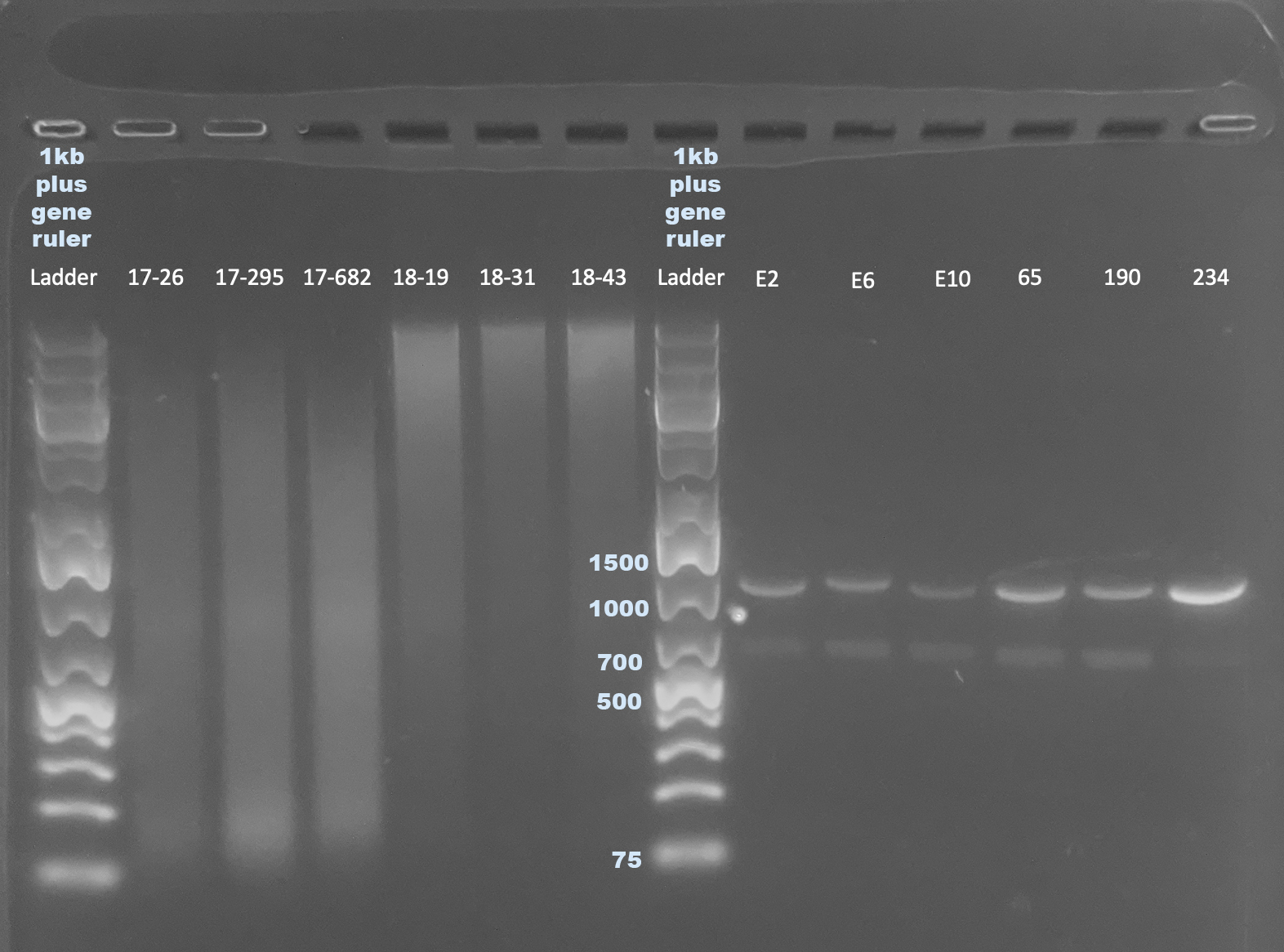
There definitely is a double-banding pattern here. It’s interesting that either band doesn’t seem to be at around 1000bp, which is the size mtORF should be. I don’t know what’s going on with that. It is also strange that the upper band seems to be around the size of that one probable contamination band, but that 234 only has that band, but was amplified in the first run where there was no contamination. We are sequencing these anyways to get an idea. Janet says there is a possibility of doing gel extractions on bands this far apart if necessary.
Written on July 31, 2020