MethylFlash Global DNA Methylation ELISA on Mcap Bleach/Non-Bleach from HI
Using the EpiGentek MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (Colormetric) on 8 Bleached and 8 Not Bleached Montipora Fragment DNA from December in HI
Goal: Learn new kit and assess global DNA methylation percentage of these 16 samples.
Results: Methylation ranges from 0.2% to 2.24% in some samples depending on how the standard curve is analyzed. All values are within the curve
Notes: Replicate samples don’t have the same or sometimes similar OD reading, including in some of the standards. This could be normal variability or it could be leftover wash buffer in some wells or uneven mixing. Additionally there are different options on how to analyze the standard curve to get the 5-mC%
MethylFlash Kit
Notes
- Samples are 16 randomly placed bleached and not-bleached Montipora from December in HI. Bleaching info has been removed from subsiquent steps
- Input to kit is ideally 100ng in 2-5ul. All samples have enough DNA for 100ng and I decided to use 5ul for each one for easy pipetting
- Each standard and sample was run in duplicate
- Kit does not say anything about what should be kept on ice or brought to room temp. I tried to use things at room temp. Except for DNA I kept on ice. The wash buffer needs to be brought to room temp before use.
- All these steps were done with filter tips
Procedure
Sample Prep
- Calculated the amount of DNA needed for 200ng (duplicate) and the volume of 10mM Tris HCl for 10ul (5 + 5)
- This is also the order the samples are in on the plate (random)
| Colony-ID | Collection-Date | Extraction-ID | DNA-ng/ul | ul for 200ng | ul 10mM Tris HCl to 10ul |
|---|---|---|---|---|---|
| M-210 | 2019-12-04 | 34 | 37.15 | 5.38 | 4.62 |
| M-3 | 2019-12-04 | 35 | 32.1 | 6.23 | 3.77 |
| M-217 | 2019-12-04 | 16 | 36.5 | 5.48 | 4.52 |
| M-12 | 2019-12-04 | 25 | 31.2 | 6.41 | 3.59 |
| M-221 | 2019-12-04 | 28 | 30.6 | 6.54 | 3.46 |
| M-20 | 2019-12-04 | 21 | 54.5 | 3.67 | 6.33 |
| M-204 | 2019-12-04 | 30 | 37.5 | 5.33 | 4.67 |
| M-218 | 2019-12-04 | 33 | 89.4 | 2.24 | 7.76 |
| M-4 | 2019-12-04 | 18 | 31.3 | 6.39 | 3.61 |
| M-219 | 2019-12-04 | 23 | 35.7 | 5.60 | 4.40 |
| M-203 | 2019-12-04 | 31 | 21.2 | 9.43 | 0.57 |
| M-222 | 2019-12-04 | 26 | 46 | 4.35 | 5.65 |
| M-211 | 2019-12-04 | 32 | 26.9 | 7.43 | 2.57 |
| M-19 | 2019-12-04 | 29 | 63.8 | 3.13 | 6.87 |
| M-220 | 2019-12-04 | 24 | 68.9 | 2.90 | 7.10 |
| M-209 | 2019-12-04 | 22 | 51.1 | 3.91 | 6.09 |
- Thawed DNA samples on ice
- Prepared strip tubes for dilutions: each sample would get 1 tube with the volume for 2 assays: I planned to take twice from the same tube to put into the plate
- Vortexed and spun down DNA
- Added Tris volume to each dilution tube
- Added DNA volume to each dilution tube
- Kept diluted DNA tubes on ice
Kit Prep
- Set incubator genie to 37 degrees
- Took all components out of the fridge, freezer, and kit box
- Made diluted wash buffer:
- 117mL of Type I DI water
- 13mL of 10X wash buffer
- Note, here I had not realized that there was precipitate in the wash buffer before beginning to add to the water. I stopped, warmed the buffer in the incubator to dissolve the precipitates, and then added the total volume in the bottle to the water. This came out to be 14.82mL, so I added and extra 16.38mL of Type I DI water to compensate.
- Swirled to mix wash buffer
- Checked wash buffer pH by using the pH meter
- Calibrated pH meter
- pH was 7.46: within the range needed: 7.2-7.5
Standard Curve Prep
- Vortexed and spun down positive (PC) and negative control (NC) (kit provided)
- Made diluted positive control (DPC)
- 9ul of NC
- 1ul of PC
- Vortex and spin down
- Made prep tubes for standards in same way as for samples: each sample would get 1 tube with the volume for 2 assays: I planned to take twice from the same tube to put into the plate
- Added 5ul of NC to NC prep tube (volume of standards to add to plate is 2ul only – this is based off using 100ng)
- 0.1% standard
- 1ul DPC
- 9ul NC
- 0.2% standard
- 1ul DPC
- 4ul NC
- 0.5% standard
- 3ul DPC
- 3ul NC
- 1% standard
- 1ul PC
- 9ul NC
- 2% standard
- 1ul PC
- 4ul NC
- 5% standard
- 3ul PC
- 3ul NC
- Vortexed and spun down all tubes and kept on ice
Actual Assay
Plate layout

- Vortexed and spun down binding solution (BS)
- Added 100ul BS to each NC well
- Added 2ul of NC to each NC well, pipetting up and down a few times
- Added 100ul BS to each standard well, 2 at a time
- Added 2ul of appropriate standard solution, 2 at a time, pipetting up and down a few times
- Added 100ul of BS to each sample well, 2 at a time
- Added 5ul of each sample to their wells, 2 at a time, pipetting up and down a few times
- Once finished, rocked the plate back and forth to make sure the bottom of the wells were covered in liquid and shook it a little gently against the table
- Covered the plate with the same seal it came with (parafilm didn’t work very well because it was only half the plate)
- Placed the plate in the incubator genie for 1 hour at 37 degrees C
- With 10 minutes left in the incubation, I made the 5mc Detection Complex Solution:
- 2.4mL diluted wash buffer
- 2.4ul mc-antibody
- Vortexed and spun down
- 2.4ul signal indicator
- 1.2ul enhancer solution
- Vortexed and spun down again
- Removed the plate from the incubator after 60min
- Pipetted out the BS into waste
- Made a basin with diluted wash buffer
- Washed each well 3 times with 150ul of diluted wash buffer:
- Used the multichannel by column
- Pipetted in and then out wash buffer one column at a time
- After the last wash, went back through and tried to suck out any remaining volume
- Also tapped the plate over a paper towel
- Then added 50ul of the 5-mc Detection Complex Solution to each well
- Covered the plate with the same seal and left at RT on the bench for 50 minutes
- After the incubation, I removed the solution by pipetting
- Washed each well 5 times with 150ul diluted wash buffer in the same way as before
- Again I tried to remove any left over wash by pipetting again after the last wash and tapping down on a paper towel
- Used a small basin for the developing solution:
- Used the multichannel to add 100ul of the developing solution to each well by column so that each replicate got solution at the same time
- Gently shook the plate against the bench
- Watched the color change in the wells for 5 minutes (the 5% standards was supposed to turn “deep blue”, I wasn’t sure what was considered deep)
- Plate after 5 min

- Used a small basin for the stop solution:
- Used the multichannel to add 100ul of the stop solution to each well by column so that each replicate got solution at the same time
- Gently shook the plate against the bench
- Waited 1-2 minutes, all the wells went from blue to yellow when adding the SS pretty much immediately
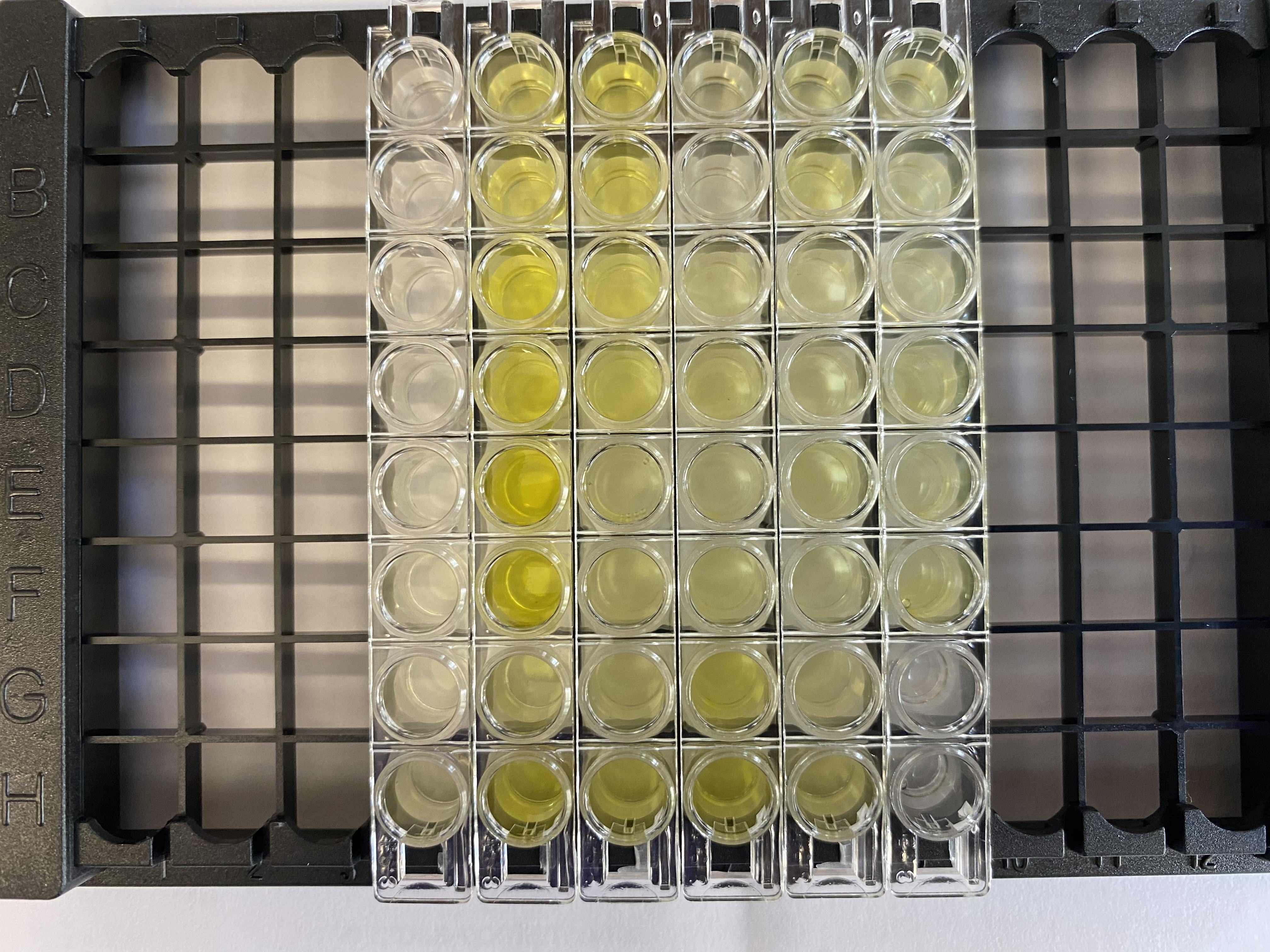
- Started up the computer and plate reader
- Read the whole plate within 15 minutes at 450nm absorbance
Results
Output from the Plate Reader in Excel Format
Raw Absorbance Values for Standard Curve
| Percent_PC | Absorbance_450 |
|---|---|
| 0 | 0.043 |
| 0 | 0.046 |
| 0.1 | 0.049 |
| 0.1 | 0.05 |
| 0.2 | 0.068 |
| 0.2 | 0.107 |
| 0.5 | 0.107 |
| 0.5 | 0.176 |
| 1 | 0.334 |
| 1 | 0.401 |
| 2 | 0.715 |
| 2 | 0.679 |
| 5 | 1.238 |
| 5 | 0.969 |
Raw Absorbance Values for Samples
| Sample | Absorbance_450 |
|---|---|
| M-210-a | 0.276 |
| M-210-b | 0.67 |
| M-3-a | 0.419 |
| M-3-b | 0.419 |
| M-217-a | 0.405 |
| M-217-b | 0.391 |
| M-12-a | 0.198 |
| M-12-b | 0.26 |
| M-221-a | 0.23 |
| M-221-b | 0.468 |
| M-20-a | 0.147 |
| M-20-b | 0.078 |
| M-204-a | 0.177 |
| M-204-b | 0.284 |
| M-218-a | 0.183 |
| M-218-b | 0.245 |
| M-4-a | 0.499 |
| M-4-b | 0.577 |
| M-219-a | 0.223 |
| M-219-b | 0.317 |
| M-203-a | 0.182 |
| M-203-b | 0.191 |
| M-222-a | 0.256 |
| M-222-b | 0.192 |
| M-211-a | 0.222 |
| M-211-b | 0.308 |
| M-19-a | 0.24 |
| M-19-b | 0.217 |
| M-220-a | 0.2 |
| M-220-b | 0.283 |
| M-209-a | 0.227 |
| M-209-b | 0.301 |
Average Absorbance Values for Samples
| Sample_ID | Average_OD |
|---|---|
| M-210 | 0.473 |
| M-3 | 0.419 |
| M-217 | 0.398 |
| M-12 | 0.229 |
| M-221 | 0.349 |
| M-20 | 0.1125 |
| M-204 | 0.2305 |
| M-218 | 0.214 |
| M-4 | 0.538 |
| M-219 | 0.27 |
| M-203 | 0.1865 |
| M-222 | 0.224 |
| M-211 | 0.265 |
| M-19 | 0.2285 |
| M-220 | 0.2415 |
| M-209 | 0.264 |
Analysis R Markdown
Input Standards csv
Input Sample OD Averages csv
Output %s using the 3 models