Using the Zymo Pico Methyl Seq Library Prep Kit for Two Final of Danielle’s Pocillopora DNA Samples for Whole Genome Bisulfite Sequencing
Goal Library prep 2 samples for WGBS that failed previously
Results Both samples worked!
Takeaways I re-diluted the sample DNA and the index primers, so I do not know if either of those was the initial problem or something else during the prep. I continue to struggle to get all preps to work on the first prep, I am not sure why. It was noticeably easier and much more manageable to do 2 samples instead of 16/17. I think that might be almost too many to do at once.
This library prep followed the exact protocol for the Zymo Pico Methyl Seq Kit from me. See that protocol for detailed descriptions of each steps. Tables and values specific for this prep are included below.
2020 10 13
Re-Dilution of Samples:
| Coral ID |
DNA for dilution to 1ng/ul (ul) |
10mM Tris HCl for dilution to 1ng/ul (ul) |
| E2 |
2 |
20.6 |
| E9 |
2 |
20.8 |
Re-Dilution of Index Primers
| Primer Name |
Volume 200uM Stock |
Volume Nuclease-free Water for 10uM |
| i5_ZM_UDI028 |
10ul |
190ul |
| i7_ZM_UDI028 |
10ul |
190ul |
| i5_ZM_UDI029 |
10ul |
190ul |
| i7_ZM_UDI029 |
10ul |
190ul |
Bisulfite Conversion
- Samples used in prep:
- Followed exact steps as in protocol
- Proceeded directly to next steps
Post-BS Conversion cleanup
- Followed exact steps as in the protocol
Amplification with Prep-Amp Primers
- Followed exact steps as in the protocol
- Priming Master Mix calculations (PMM):
- 2ul PrepAmp Buffer * 2.2 = 34.4ul
- 1ul PrepAmp Primer * 2.2 = 2.2ul
- PrepAmp Master Mix calculations (PAMM):
- 1ul PrepAmp Buffer * 2.2 = 2.2ul
- 3.75ul PrepAmp PreMix * 2.2 = 8.25ul
- 0.3ul PrepAmp Polymerase * 2.2 = 0.66ul
- Dilution calculation of PrepAmp Polymerase to add 0.5ul:
- 0.3 PrepAmp Polymerase * 2.2 = 0.66ul
- 0.2ul DNA elution buffer * 2.2 = 0.34ul
- Note: it was very hard to pipette 0.66, this may have been slightly off
DNA Clean and Concentrator
- Followed exact steps as in the protocol
First Amplification
- Followed exact steps as in the protocol
- 1st Amp Master Mix calculation:
- 12.5ul Library Amp Mix * 2.2 = 27.5ul
- 1ul Library Amp Primers * 2.2 = 2.2ul
Second DNA Clean and Concentrator
- Followed exact steps as in the protocol
Second Amplification with Index Primers
- Followed exact steps as in the protocol, except see below
- Table for components in tubes for amplifications:
| Sample |
Volume DNA (ul) |
Volume Library Amp Mix (ul) |
Volume i5 Primer (10uM) |
Volume i7 Primer (10uM) |
| E2 |
12 |
14 |
1ul i5_ZM_UDI028 |
1ul i7_ZM_UDI028 |
| E9 |
12 |
14 |
1ul i5_ZM_UDI029 |
1ul i7_ZM_UDI029 |
1X Bead Clean
- Followed exact steps as in protocol
- Samples were Qubited immediately so they were put on an ice bucket not frozen yet
Broad Range dsDNA Qubit
| Sample |
Reading 1 (ng/ul) |
Reading 2(ng/ul) |
Average (ng/ul) |
| Standard 1 |
181 RFU |
- |
- |
| Standard 2 |
19823 RFU |
- |
- |
| E2 |
13.8 |
13.7 |
13.75 |
| E9 |
15.4 |
15.3 |
15.35 |
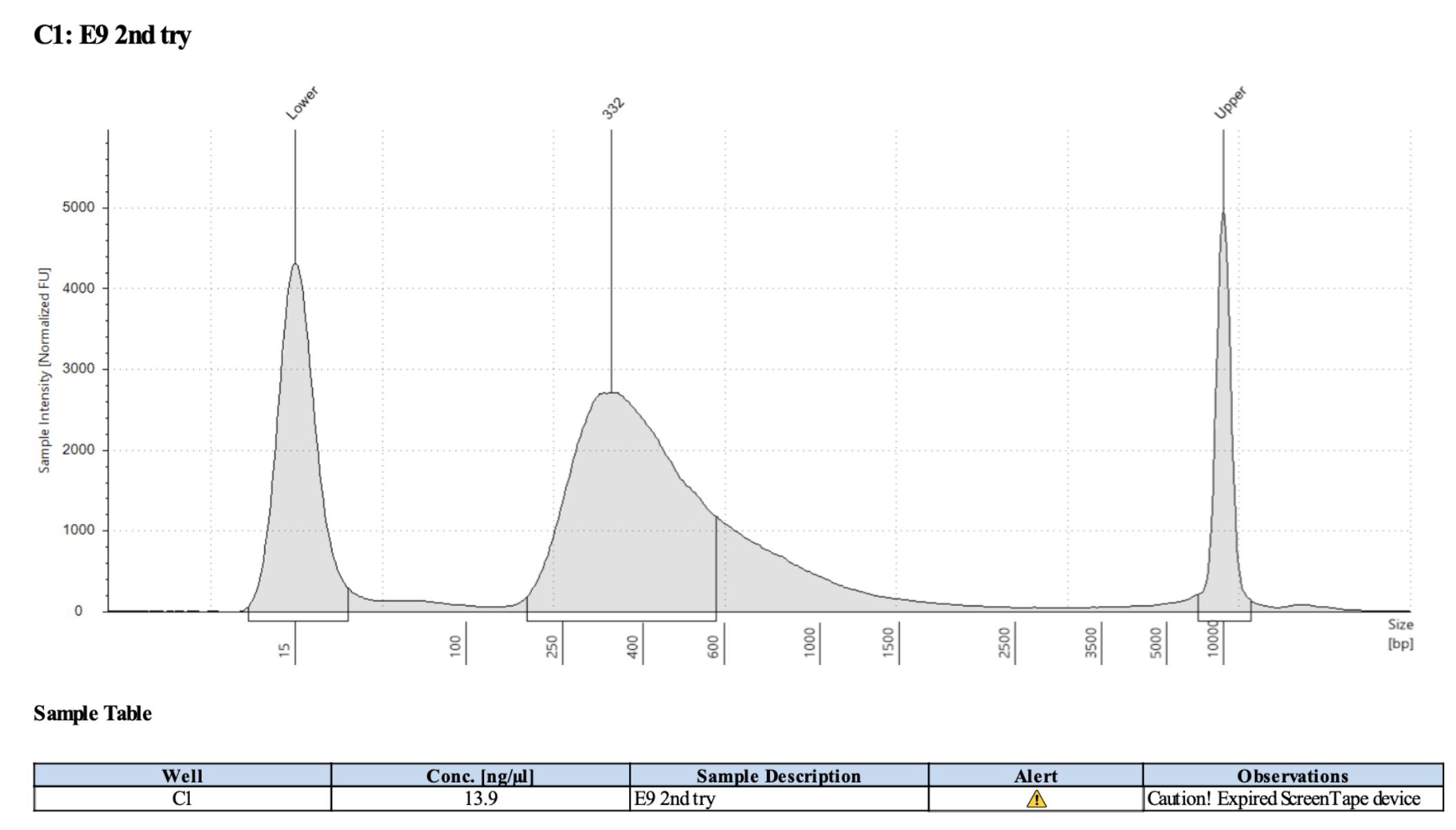
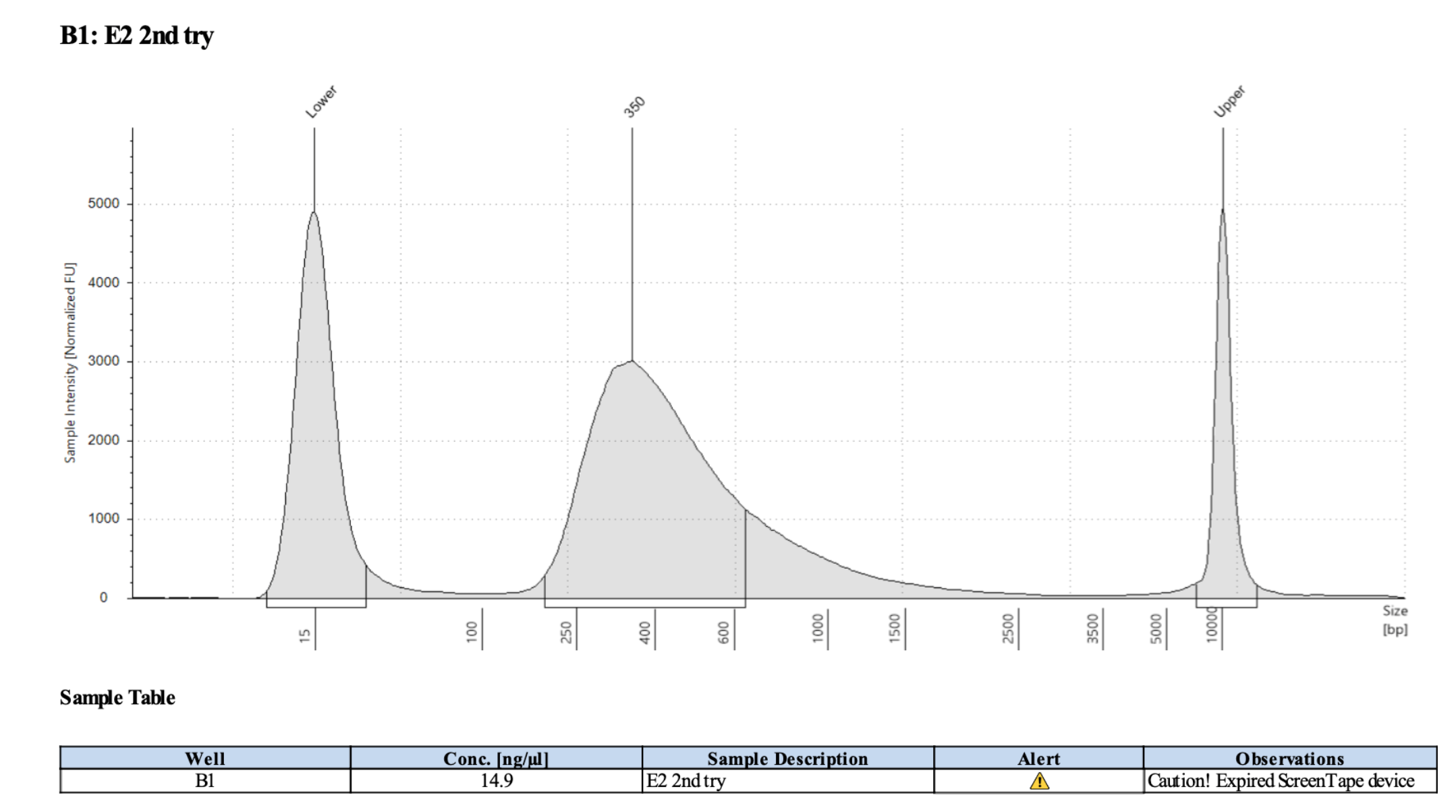
D5000 TapeStation
Samples and Index Sequences
| Coral ID |
i7 bases |
i5 bases |
| E2 |
TCCAACGC |
TTGGACTT |
| E9 |
CCGTGAAG |
CAGTGGAT |
 E9 Library:
E9 Library: