Pocillopora Samples from Danielle mtORF Amplification and Sanger Sequencing Prep Processing
mtORF Amplification and Sanger Sequencing Prep for Danielle’s Pocillopora Samples
Goal: Amplify Danielle’s 32 samples, with 3 control reactions for mtORF. Then dilute them properly and send them to the GSC for Sanger sequencing.
Major Results: Amplifications were really great this time, there was one clear band at ~1000bp which is exactly what we’re supposed to see. It was interesting that rows of the plate had more similar post-PCR concentrations to each other than random samples, even though I tried to dilute all the samples before hand to close to the same concentration.
Major Takeaways: The protocol seems to be working great when scaled up to more samples than the test. I thought about if it would be better to increase the elution volume from the beads so you don’t have to dilute before preparing to sequence, but it would be not good if the concentration then became too low to pipette 25ng in 10ul, and because the PCR concentrations were more variable than I thought they would, I am not decided on if increasing the elution volume from 50ul to like 70ul is a good idea or not. You can always dilute but concentrating is a hassle.
Process
Followed the mtORF amplification protocol exactly. See that for in depth details for this protocol
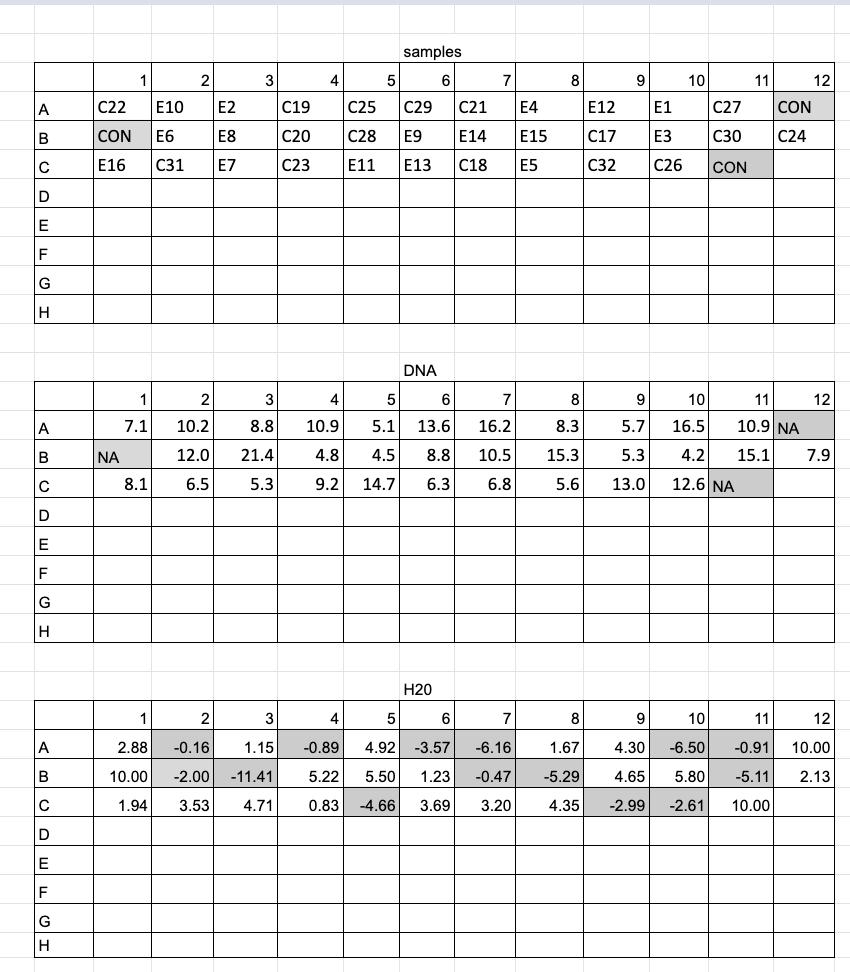
DNA Dilutions 2020-08-27
- Arranged samples in a 96 well plate randomly with 3 spots for control reactions
- Allocated DNA for 10ng/ul in 10ul of nuclease free water, so 100ng of DNA. If samples had concentrations below 10ng/ul to start with, just 10ul of those samples was added to each well
mtORF Amplifications 2020-08-27
- 32 samples plus three negative controls is 35 reactions, use an additional 3 for error
- Made a master mix for 38 samples and 3 reactions each:
- 1900ul Phusion master mix
- 49.4ul FatP6.1 10uM Primer
- 49.4ul RORF 10uM Primer
- 1672ul nuclease free water
- Added 97ul of master mix into 35 wells in a new plate
- Used a multichannel to add 3ul of DNA from the dilution plate in to the same orientation wells in the plate with the master mix
- Covered plate and vortexed and spun down
- Separated plate out 2 times into 3 separated reaction mixes each with 33ul
- Covered plates, spun down, and placed in three thermocyclers FatP6.1 RORF program
1X Bead Cleanup and Quantification 2020-08-28
- Combined triplicate reactions back together
- Added 1X (100ul) beads to each well
- Followed bead cleanup protocol
- Resuspended and eluted DNA in 50ul ultra-pure water and removed into a new plate (same orientation)
- dsDNA broad range Qubit assay for 35 samples (n# 40)
| Sample.ID | Qubit reading 1 (ng/ul) | Qubit reading 2 (ng/ul) | average DNA (ng/ul) |
|---|---|---|---|
| C22 | 29.6 | 30.2 | 29.9 |
| E10 | 30 | 30 | 30 |
| E2 | 27 | 26.6 | 26.8 |
| C19 | 25.8 | 25.8 | 25.8 |
| C25 | 27 | 26.8 | 26.9 |
| C29 | 22 | 21.8 | 21.9 |
| C21 | 25.2 | 25.2 | 25.2 |
| E4 | 22.2 | 22 | 22.1 |
| E12 | 28 | 28.2 | 28.1 |
| E1 | 23 | 22.4 | 22.7 |
| C27 | 21.8 | 22 | 21.9 |
| E6 | 23 | 23.2 | 23.1 |
| E8 | 21.8 | 21.8 | 21.8 |
| C20 | 23 | 23.2 | 23.1 |
| C28 | 21.6 | 21.6 | 21.6 |
| E9 | 23.2 | 23.4 | 23.3 |
| E14 | 17.7 | 17.7 | 17.7 |
| E15 | 17.5 | 17.6 | 17.55 |
| C17 | 21 | 21 | 21 |
| E3 | 21.8 | 21.8 | 21.8 |
| C30 | 18.4 | 18.4 | 18.4 |
| C24 | 19.3 | 19.4 | 19.35 |
| E16 | 39.4 | 39.8 | 39.6 |
| C31 | 35.2 | 35.4 | 35.3 |
| E7 | 42 | 42.4 | 42.2 |
| C23 | 38 | 38.4 | 38.2 |
| E11 | 44.2 | 44.4 | 44.3 |
| E13 | 45.4 | 45.6 | 45.5 |
| C18 | 42.8 | 43.2 | 43 |
| E5 | 46 | 45.6 | 45.8 |
| C32 | 38 | 38.2 | 38.1 |
| C26 | 37.8 | 37.8 | 37.8 |
1% Gel To Confirm Bands 2020-09-01
-
Made a 1% gel in the medium gel box

-
One band at ~1000bp, exactly right! No contamination, only imaging artifacts
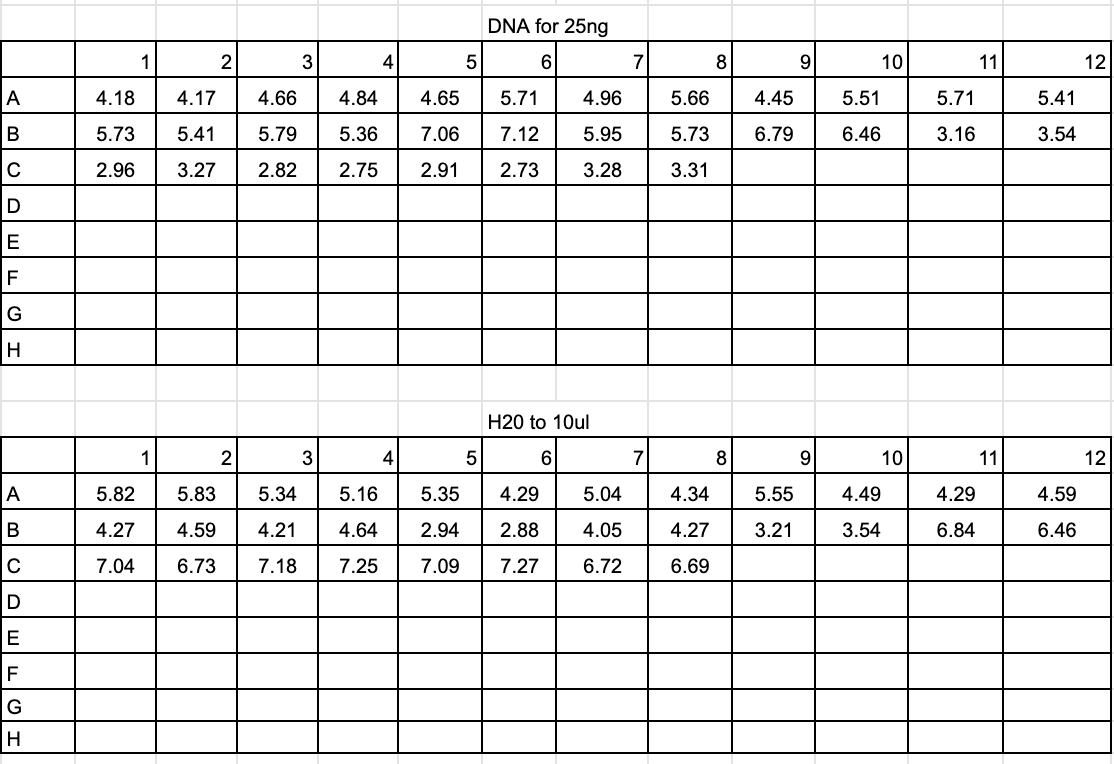
Dilution and Sequencing Prep 2020-09-02
- Diluted each DNA amplification by 1:5 (2ul of DNA and 10ul of ultra-pure water)
- Created plate with 25ng of DNA for each sample and ultra-pure water up to 10ul
- Added 2ul of 3.2uM RORF primer to each well
- Covered, vortexed, spun down plate
- Brought upstairs for sequencing the next day
Spreadsheet for Sequencing
| Sample ID (lab) | Sample IDa (GSC) | Well (GSC use only) | Template Typeb | A. Template Size (bases) | B. Template Stock Conc. (ng/µl) | C. PCR template: ng needed = ((A ÷ 100) x 1.25) x 2 | D. PCR template: Volume = (C ÷ B) µl | F. Volume PCR-H20 needed (10 minus D or E) µl | G. Volume primer needed 1 µl per reaction |
|---|---|---|---|---|---|---|---|---|---|
| C22 | HPD1 | PCR | 1000 | 5.98 | 25 | 4.18 | 5.82 | 2 | |
| E10 | HPD2 | PCR | 1000 | 6 | 25 | 4.17 | 5.83 | 2 | |
| E2 | HPD3 | PCR | 1000 | 5.36 | 25 | 4.66 | 5.34 | 2 | |
| C19 | HPD4 | PCR | 1000 | 5.16 | 25 | 4.84 | 5.16 | 2 | |
| C25 | HPD5 | PCR | 1000 | 5.38 | 25 | 4.65 | 5.35 | 2 | |
| C29 | HPD6 | PCR | 1000 | 4.38 | 25 | 5.71 | 4.29 | 2 | |
| C21 | HPD7 | PCR | 1000 | 5.04 | 25 | 4.96 | 5.04 | 2 | |
| E4 | HPD8 | PCR | 1000 | 4.42 | 25 | 5.66 | 4.34 | 2 | |
| E12 | HPD9 | PCR | 1000 | 5.62 | 25 | 4.45 | 5.55 | 2 | |
| E1 | HPD10 | PCR | 1000 | 4.54 | 25 | 5.51 | 4.49 | 2 | |
| C27 | HPD11 | PCR | 1000 | 4.38 | 25 | 5.71 | 4.29 | 2 | |
| E6 | HPD12 | PCR | 1000 | 4.62 | 25 | 5.41 | 4.59 | 2 | |
| E8 | HPD13 | PCR | 1000 | 4.36 | 25 | 5.73 | 4.27 | 2 | |
| C20 | HPD14 | PCR | 1000 | 4.62 | 25 | 5.41 | 4.59 | 2 | |
| C28 | HPD15 | PCR | 1000 | 4.32 | 25 | 5.79 | 4.21 | 2 | |
| E9 | HPD16 | PCR | 1000 | 4.66 | 25 | 5.36 | 4.64 | 2 | |
| E14 | HPD17 | PCR | 1000 | 3.54 | 25 | 7.06 | 2.94 | 2 | |
| E15 | HPD18 | PCR | 1000 | 3.51 | 25 | 7.12 | 2.88 | 2 | |
| C17 | HPD19 | PCR | 1000 | 4.2 | 25 | 5.95 | 4.05 | 2 | |
| E3 | HPD20 | PCR | 1000 | 4.36 | 25 | 5.73 | 4.27 | 2 | |
| C30 | HPD21 | PCR | 1000 | 3.68 | 25 | 6.79 | 3.21 | 2 | |
| C24 | HPD22 | PCR | 1000 | 3.87 | 25 | 6.46 | 3.54 | 2 | |
| E16 | HPD23 | PCR | 1000 | 7.92 | 25 | 3.16 | 6.84 | 2 | |
| C31 | HPD24 | PCR | 1000 | 7.06 | 25 | 3.54 | 6.46 | 2 | |
| E7 | HPD25 | PCR | 1000 | 8.44 | 25 | 2.96 | 7.04 | 2 | |
| C23 | HPD26 | PCR | 1000 | 7.64 | 25 | 3.27 | 6.73 | 2 | |
| E11 | HPD27 | PCR | 1000 | 8.86 | 25 | 2.82 | 7.18 | 2 | |
| E13 | HPD28 | PCR | 1000 | 9.1 | 25 | 2.75 | 7.25 | 2 | |
| C18 | HPD29 | PCR | 1000 | 8.6 | 25 | 2.91 | 7.09 | 2 | |
| E5 | HPD30 | PCR | 1000 | 9.16 | 25 | 2.73 | 7.27 | 2 | |
| C32 | HPD31 | PCR | 1000 | 7.62 | 25 | 3.28 | 6.72 | 2 | |
| C26 | HPD32 | PCR | 1000 | 7.56 | 25 | 3.31 | 6.69 | 2 |