Second Qiagen Genomic Tip for HMW DNA Extraction of Porites asteroides
Second Attempt At High Molecular Weight DNA Extraction of Porites asteroides with the Qiagen Genomic Tip for Intended Genome Sequencing
In this protocol I use the QIAGEN Genomic-tip 100/G, the QIAGEN Genomic DNA Buffer Set, QIAGEN RNase A (100mg/mL concentration), QIAGEN Proteinase K, and DNA lo-bind tubes
Previous attempt for this extraction did not yield enough DNA, so I decided to chisel more weight to see if a lack of sample was one of the major hold ups. Additionally DNA lo-bind tubes were used to prevent sticking of DNA to the tubes.
Goal: Get 25ug of HMW DNA from this sample
Result: 39ug of DNA
Major Take Aways: For this species, much more than the recommended amount of tissue is needed. Additionally, even after all steps were taken to prevent clogging of the resin column, liquid still went slowly though it.
Sample Chiseling
Set up
- Prepared digestion buffer with 9.5mL of Buffer G2 and 19ul of RNase A
- Set incubator genie to 50 degrees C
- Cleaned chisel, forceps, scraper/scooper and bench with 10% bleach, DI water, then 70% EtOH
- Wrapped those in foil and placed in -80
- Also placed mortar and pestle in -80
- Had styrofoam cooler of dry ice saved from a shipment and filled Thermoflask dewar with LN2
- Brought over scale
- Set up dry ice box with plastic tube box rack over the ice to have a hard surface for chiseling, then covered that in foil
Chiseling
- Kevin Wong helped by holding the nubbin steady at the base
- Took out nubbin: TA HOT 1 and unwrapped from foil (sample was flash frozen and kept at -80)
- Chiseled with large chisel and hammer, increased hammering strength considerably this time and large chunks came off
- Took mortar from -80 on the scale and tarred
- Used forceps to put chunks into mortar: 0.47g
- Poured LN2 into mortar and let boil off
- Ground chunks with pestle until powdery, image:

- Scrapped into a chilled 50mL conical with the spatula
- Poured in the buffer G2 mix
- Vortexed briefly
- Added 500ul of Proteinase K and vortexed again
- Image of liquid before incubation:

- Placed in incubator genie at 50 degrees C for 2 hours, rocking at 15 speed
Genomic Tip Extraction
Genomic Tip
- Set centrifuge to 4 degrees C
- Image of liquid after incubation:

- After incubation, transferred 1mL of lysed tissue liquid into each of 10 1.5mL tubes with wide bore pipette tips
- Centrifuged at 4 degrees C for 10 minutes at 5000 rcf
- Set up tip (resin column) inside a holder over a 50mL conical
- While that was going, added 4mL of equilibration buffer (QBT) to the tip and let drip through to the conical (took the 10 min)
- After centrifugation sample image:
- Added the supernatant from the sample tubes to the resin tip with wide bore pipette tip and let drip through
- Dripping took ~35 minutes
- Changed 50mL waste conicals
- Added 7.5mL of buffer QC (wash) and let drip through (30min)
- Pipetted 5mL of buffer QF into a 5mL tube and warmed in incubator genie to 50 degrees C
- Repeated wash addition (30 minutes)
- Transferred resin tip to a different 50mL conical
- Added the 5mL of warmed buffer QF and let drip through (25 minutes)
Isopropanol Precipitation of DNA
- Made 7 DNA lo-bind 1.5mL tubes, 6 each with 833ul of the elution liquid, the last tube was about 250ul
- Added 583ul (0.7 volumes) of room temp 100% isopropanol to each of the first 6 tubes and 175ul to the 7th tube
- Gently inverted to mix
- I noticed a DNA clump form after mixing! Image:
- Centrifuged all lo-bind tubes at 10,000 rcf for 30 minutes at 1 degree C
- Made fresh 70% EtOH and placed in -20 freezer to cool down
- Once finished, looked for pellets: there was a visible brownish pellet in each tube
- One tube at a time, removed most of the supernatant
- One tube at a time, added 1mL of cold 70% EtOH and vortexed briefly
- Centrifuged tubes for 10 minutes at 4 degrees C 10,000 rcf
- Removed all of supernatant when finished, used a p200 to get small volumes out
- Let tubes air dry ~7 minutes
- Added 50ul of TE buffer to each of the 70 tubes very gently
- Incubated for 1hr at 55 degrees C in the Theremomixer
- Once done, transferred to a shaker overnight 300rpm
QC
- Broad Range Qubit next day (protocol)
- Flicked tube and took from both top T and bottom B of the tube measurements
| Standard 1 | Standard 2 | Sample | Average DNA ng/µl | Averaged top and bottom ng/ul |
|---|---|---|---|---|
| 161 RFU | 22171 RFU | 7 T | 64.1 | |
| - | - | 7 B | 255 | 159.55 |
| - | - | 8 T | 111 | |
| - | - | 8 B | 206 | 158.5 |
| - | - | 9 T | 99.8 | |
| - | - | 9 B | 115.5 | 105.3 |
| - | - | 10 T | 95.1 | |
| - | - | 10 B | 76.2 | 85.65 |
| - | - | 11 T | 82.6 | |
| - | - | 11 B | 224 | 153.3 |
| - | - | 12 T | 121.5 | |
| - | - | 12 B | 126 | 123.75 |
| - | - | 13 T | 41.2 | |
| - | - | 13 B | 42.3 | 41.75 |
- Genomic DNA screentape (protocol)
Note that in this TapeStation run, the program did not pick up the highest peak in the ladder (48000bp) so it’s size quantification ability is a little skewed.
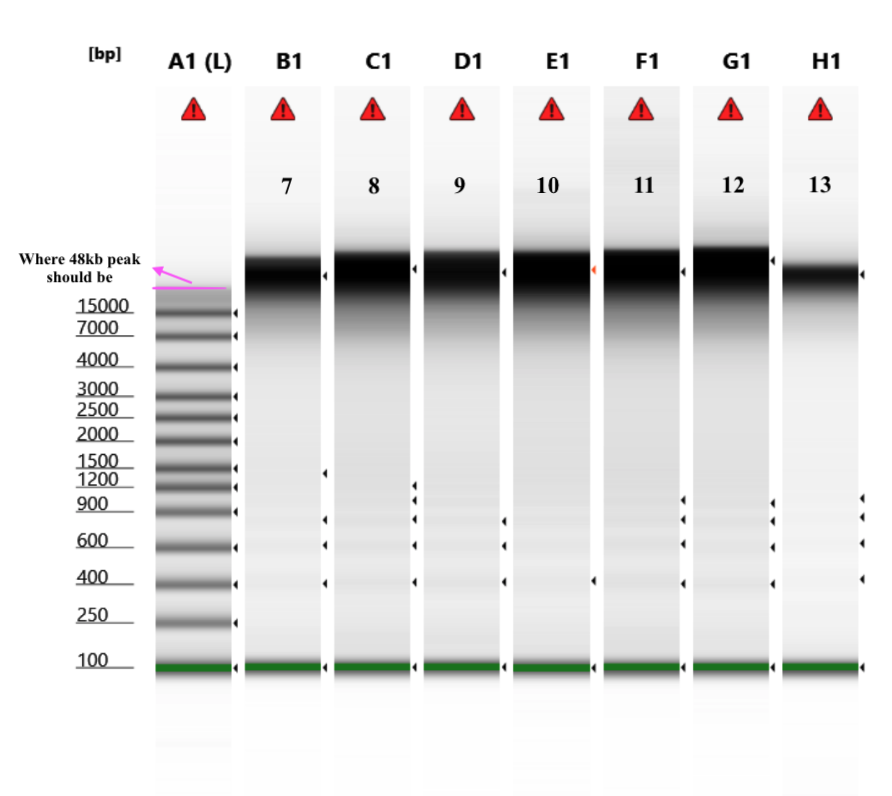
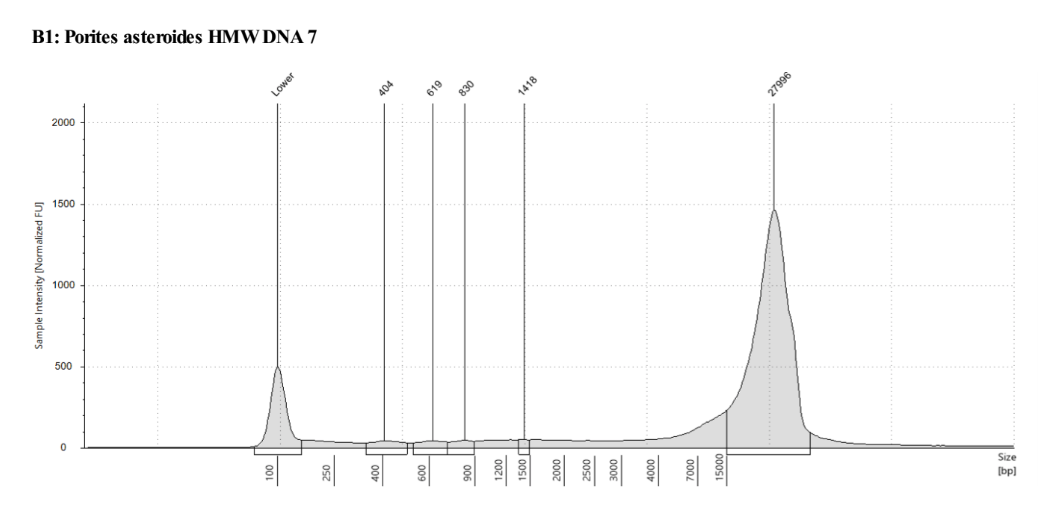
Total Amount of DNA from this extraction based on average top and bottom Qubit values and ~47ul of sample volume in each tube
- 7: 7498.85ng DNA
- 8: 7449.5ng DNA
- 9: 4949.1ng DNA
- 10: 4025.55ng DNA
- 11: 7205.1ng DNA
- 12: 5816.25ng DNA
- 13: 1962.25ng DNA
- TOTAL: 38906.5ng DNA or 38.9ug of DNA