Enriching for highly methylated regions with DNA samples of Eastern Oyster from Rebecca Stevick using the Invitrogen Methylminer Methylated DNA Enrichment Kit
Shearing DNA
DNA from Rebecca is 1µg in 80µl, 80µl is the input for the kit so it was sheared in that volume. Shearing time was 1 minute and 30 seconds, 15 seconds on 15 seconds off, at 25% amplitude, because in other methylminer preps this is about the length of time that gave a distribution of fragments between 400-600bp.
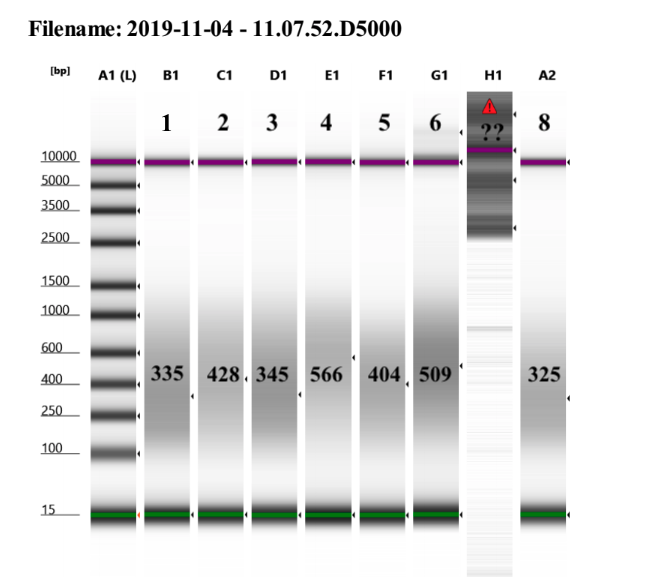
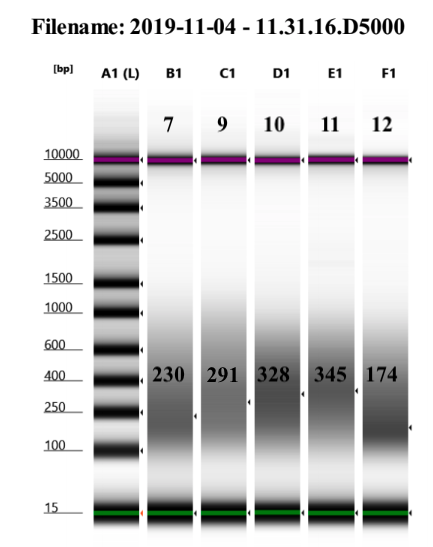
However, these were very variable. This could be because I did not check the DNA before shearing, and the previous shearing at this time was for corals. Yet, we decided to proceed because the kit only specifies bp under 1000bp, which we have, and the Pico Methyl Seq kit which I am to use for library prep next does not have an insert size requirement as it uses random priming to make the libraries.
First step was to write out the steps for the day so I would know how much 1X Bind/Wash Buffer to dilute, as it comes in 5X in the kit, that way it was ready when it was needed. 10mL made. NOTE: I had planned on doing all 12 samples, however there was not enough beads/MBD protein and then I realized the kit is for 25 samples not 30 so I ended up only doing 9. However I had made enough wash buffer for 12 so I saved it for the next day (because they give you more wash buffer proportionally than other reagents)
Preparing Beads
- Pipetted up and down the Dynabeads M-280 Streptavidin to resuspend them
- Made 9 1.5mL tubes, each with 10µl of Dynabeads (the amount recommended for less than or equal to 1µg of DNA input)
- Brought volume up to a total of 100µl with 90µl of 1X bind/wash buffer to each tube
- Pipetted to mix
- Placed tubes on long magnet rack and removed and discarded supernatant when clear
- Removed tubes from rack and resuspended beads in 100µl 1X bind/wash buffer
- Placed tubes on long magnet rack and removed and discarded supernatant when clear
- Removed tubes from rack and resuspended beads in 100µl 1X bind/wash buffer
Coupling MBD-Biotin Protein to the Beads
- Thawed MBD-Biotin protein from -80 on ice
- Made 9 new 1.5mL tubes, each with 7µl of MBD-Biotin protein (amount recommended for 1µg of DNA input)
- Added 93µl of 1X bind/wash buffer to each tube to get up to a total of 100µl
- Transferred diluted protein to the washed bead tubes for a total volume of 200µl in each of 2 tubes
- Put samples on the rotisserie mixer for 1 hour at room temp
Washing MBD-Biotin-Coupled Beads
- Spun down briefly tubes from above
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Repeated: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Repeated again: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
- Mixed beads on rotisserie mixer for 5 minutes at room temp
- Finally: Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed supernatant when clear
- Resuspended beads in 100µl 1X bind/wash buffer and pipetted to mix
Capture Reaction
- To 9 new labeled 1-9 1.5mL tubes, added 20µl each of 5X bind/wash buffer
- To the appropriate tube, added the 79µl of sheared DNA and pipetted to mix
- Transferred all of each diluted DNA sample to separate tubes with the MBD-Biotin bound beads for a total of 200µl in each tube and pipetted to mix
- Mixed on rotisserie mixer overnight at 4 degrees C in the cold room
Nov 5th 2019, Removing Non-Captured DNA
- Again started by writing out protocol to calculate how much 1X bind/wash buffer to dilute (3,600µl needed)
- Took tubes out of the cold room rotisserie and spun down briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in tubes labeled non-captured DNA and sample number
- Resuspended beads in 200µl of 1X bind/wash buffer and placed on rotisserie mixer for 3 minutes
- Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in tubes labeled wash and the sample number
- Repeated: added 200µl 1X bind/wash buffer and placed on rotisserie mixer for 3 minutes
- Spun down tubes briefly
- Placed tubes on magnet rack for 1 minute
- Removed clear supernatant and SAVED in the same tubes labeled wash and the sample number for a total of 400µl in each tube
Single Fraction Elution
- Resuspended beads in 200µl High Salt Elution buffer
- Incubated on rotisserie mixer for 3 minutes
- Spun down tubes breifly and placed on magnet rack for 1 minute
- Removed supernatant when clear and SAVED in a new 1.5mL tube labeled captured DNA and the sample number
- Repeated: added 200µl High Salt Elution Buffer to resuspend beads in each tube
- Incubated tubes on rotisserie mixer for 3 minutes
- Spun down tubes briefly and placed on magnet rack for 1 minute
- Removed clear supernatant and SAVED in each sample tube for captured DNA, for a total of 400µl in each tube
Ethanol Precipitation
- Each Captured fraction tube got:
- 40µl 3M sodium acetate pH 5.2
- 800µl 100% EtOH
- 1µl glycogen
- Vortexed to mix and spun down
- Placed tubes in the -80 freezer for 2 hours
- Set centrifuge to 4 degrees C (ours can actually get to 1 degree so next time do that) with about 15 min time left and let it run to get down to temp
- Prepared 4mL of 70% EtOH and put in -20 to chill down
- Took sample tubes out of -80 and centrifuged for 15 minutes at 4 degrees C and 14,000 rcf
- Took tubes out of centrifuge one at a time to keep cold, looked at tubes to see if there was a pellet. There seemed to be a pellet in all samples, YAY
- Removed the supernatant and discarded very carefully
- Added 500µl of cold 70% EtOH to each tube carefully
- Centrifuged for 5 minutes at 4 degrees C and 14,000 rcf
- Very carefully removed supernatant from pellet
- Centrifuged tubes for 5 minutes at 4 degrees C at 14,000 rcf
- Removed any remaining supernatant as best as possible with the smallest pipette tip and making sure to not removed the pellet
- Air dried pellet for 3 minutes
- Resuspended pellets in 25µl of ultra pure water
High Sensitivity DNA Qubit
- Followed Qubit protocol for HS reagents
| Sample | Standard 1 (RFU) | Standard 2 (RFU) | 1st reading (ng/ul) | Second reading (ng/ul) | Average ng/ul |
|---|---|---|---|---|---|
| 1 Cap | 45.95 | 24944 | 2.36 | 2.34 | 2.35 |
| 2 Cap | 45.95 | 24944 | 2.52 | 2.52 | 2.52 |
| 3 Cap | 45.95 | 24944 | 1.81 | 1.8 | 1.8 |
| 4 Cap | 45.95 | 24944 | 2.36 | 2.36 | 2.36 |
| 5 Cap | 45.95 | 24944 | 1.76 | 1.76 | 1.76 |
| 6 Cap | 45.95 | 24944 | 2.34 | 2.34 | 2.34 |
| 7 Cap | 45.95 | 24944 | 1.4 | 1.4 | 1.4 |
| 8 Cap | 45.95 | 24944 | 0.836 | 0.83 | 0.833 |
| 9 Cap | 45.95 | 24944 | 1.98 | 1.98 | 1.98 |