Puregene DNA Extraction of Same Dinn Cells Infected with DiNV That Were Used to Test The Hirt Extraction, PCRs of All Samples, and Qubit
20231115 Puregene Extraction
If I want to see if the Hirt extraction method is doing anything to increase the ratio of DiNV DNA to cell DNA, I need to compare it to samples extracted with our regular Puregene method. When harvesting the samples, I took 50ul from each flask of the supernatant/cell mix and froze that. I used those samples, labeled as A and B, for the Puregene extraction.
All samples were thawed on ice, this general protocol was used, and filter tips were used at all times. Since all samples were presumed positive for DiNV, there was no specific order to them. 2 Extraction controls were done at the same time as well.
PCRs 20231116
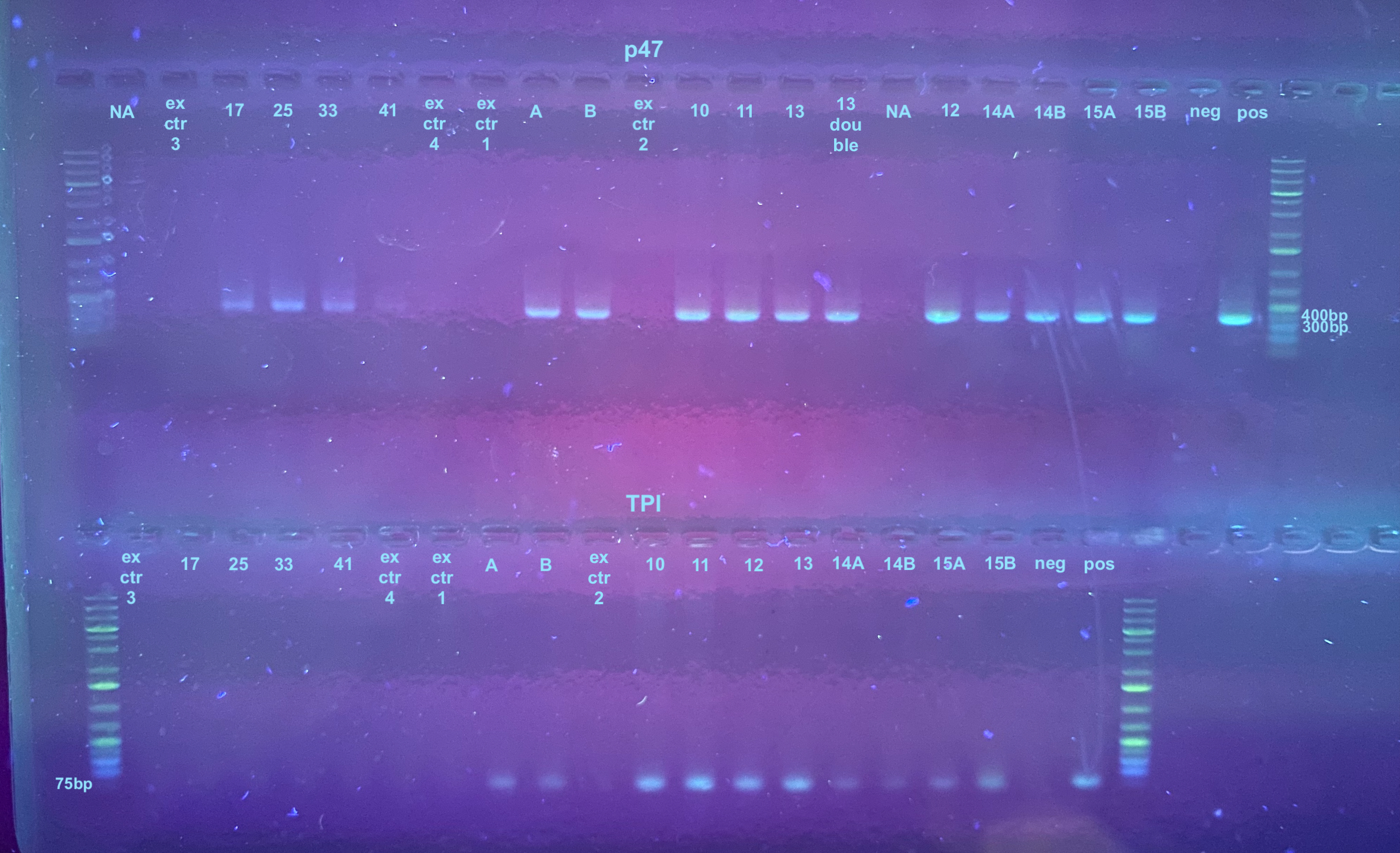
The extraction controls, samples A and B, and the 8 samples from the Hirt Extraction were all checked with a p47 and TPI PCR. nformation on the primers and their programs can be found here, but the TPI program had an extension time of 30 seconds.
The PCR process followed this general protocol. Afterwards a 1% gel was run for 25 minutes at 100V. There are a few other samples on this gel that I was running at the same time. Samples start at ex ctr 1 and to the right of that.
Qubit 20231116
I wanted to quantify these samples to potentially use them for qPCR. I also plan on adding in some infected fly samples for the qPCR, so I quantified those as well. I followed the Qubit protocol.
- A: too low
- B: too low
- 10: 73.4ng/ul
- 11: 69.5ng/ul
- 12: 34.3ng/ul
- 13: 27.5ng/ul
- 14A: 2.58ng/ul
- 15A: too low
- fly 14: 19.2
- fly 15: 18.7
- fly 17: 9.51
- fly 46: 17.6
- fly 47: 5.17
- fly 62: 2.46
- fly 65: 14