DNA Extractions and CO1 and p47 PCRs of Day -6 of DiNV Experimental Evolution Samples
There are 3 samples (replicates of the same flask) that I need to DNA extract and PCR to check to see that these cells are virus negative.
Note for this DNA extraction the centrifuge was in the fridge and I did not move it, so all spins are at 4C.
| tube number | day sampled | sample volume |
|---|---|---|
| 1 | day -6 | 50ul |
| 2 | day -6 | 50ul |
| 3 | day -6 | 50ul |
- Thawed samples on ice
- Chilled cell lysis buffer on ice until cloudy
- Turned on heat blocks to 37C and 65C
- Added 250ul of cold cell lysis solution to each sample tube and pipette mixed 10X
- Incubated the tubes in the 65C heat block for 10 minutes
- Let tubes get to room temperature
- Prepared 1mg/mL RNase A from 10mg/mL
- 9ul molec grade water
- 1ul 10mg/mL RNase A
- Added 2ul of diluted RNase A to each tube
- Inverted tubes 25X to mix
- Incubated tubes on the 37C heat block for 30 minutes
- Prepared fresh 70% ethanol and 100% isopropanol
- Labeled 3 new tubes to be the final tubes
- Added 100ul 100% isopropanol to those final tubes
- After the 30 minute incubation, took the tubes out and let them come to room temperature
- Added 100ul of protein precipitation solution to each tube
- Vortexed the tubes for ~5 seconds
- Placed the tubes on ice for 5 minutes
- Centrifuged the tube max speed for 3 minutes
- Transferred the supernatant to the new tubes containing the 100% isopropanol (~400ul)
- Inverted the tubes 50X
- Centrifuged the tubes max speed 5 minutes
- Discarded the supernatant (there was no visible pellet in any tube but that is normal for cell DNA extractions)
- Added 300ul of fresh 70% ethanol to each tube
- Inverted the tubes twice
- Centrifuged the tubes max speed for 5 minutes
- Discarded the supernatant
- Let the tubes air dry for an hour
- Resuspended the DNA in 20ul of DNA hydration solution
- Usually here the DNA is left to sit for ~overnight, but I went directly into doing the PCRs
CO1 and p47 PCRs
- A positive and negative control was used for each primer. The positive control was sample 80 which was previously positive for virus
- All samples, reagents, and primers were thawed on ice, kept on ice, and vortexed and spun down before use
- Master mixes were made on ice
- p47 master mix:
- 27.5ul GoTaq
- 1.375ul p47 F
- 1.375ul p47 R
- 19.25ul molec grade water
- CO1 master mix:
- 27.5ul GoTaq
- 1.375ul CO1 F
- 1.375ul CO1 R
- 19.25ul molec grade water
- 9ul of master mixes were pipetted into strip tubes, 5 tubes per mix
- 1ul of DNA was pipetted into their respective tube. The negative control tube got 1ul of molecular grade water
- The PCR tubes were vortexed and spun down before use
- Tubes were placed in their respective thermocycler programs (see here), and p47 was run for 30 cycles
20230915 A 0.7% gel was run (we are trying to cut down on the amount of agarose used and this should work just as well as a 1%), with 40mL of 1X TAE, 0.35g of agarose, and 0.7ul of Midori stain. The gel was run at 90V for 30 minutes:
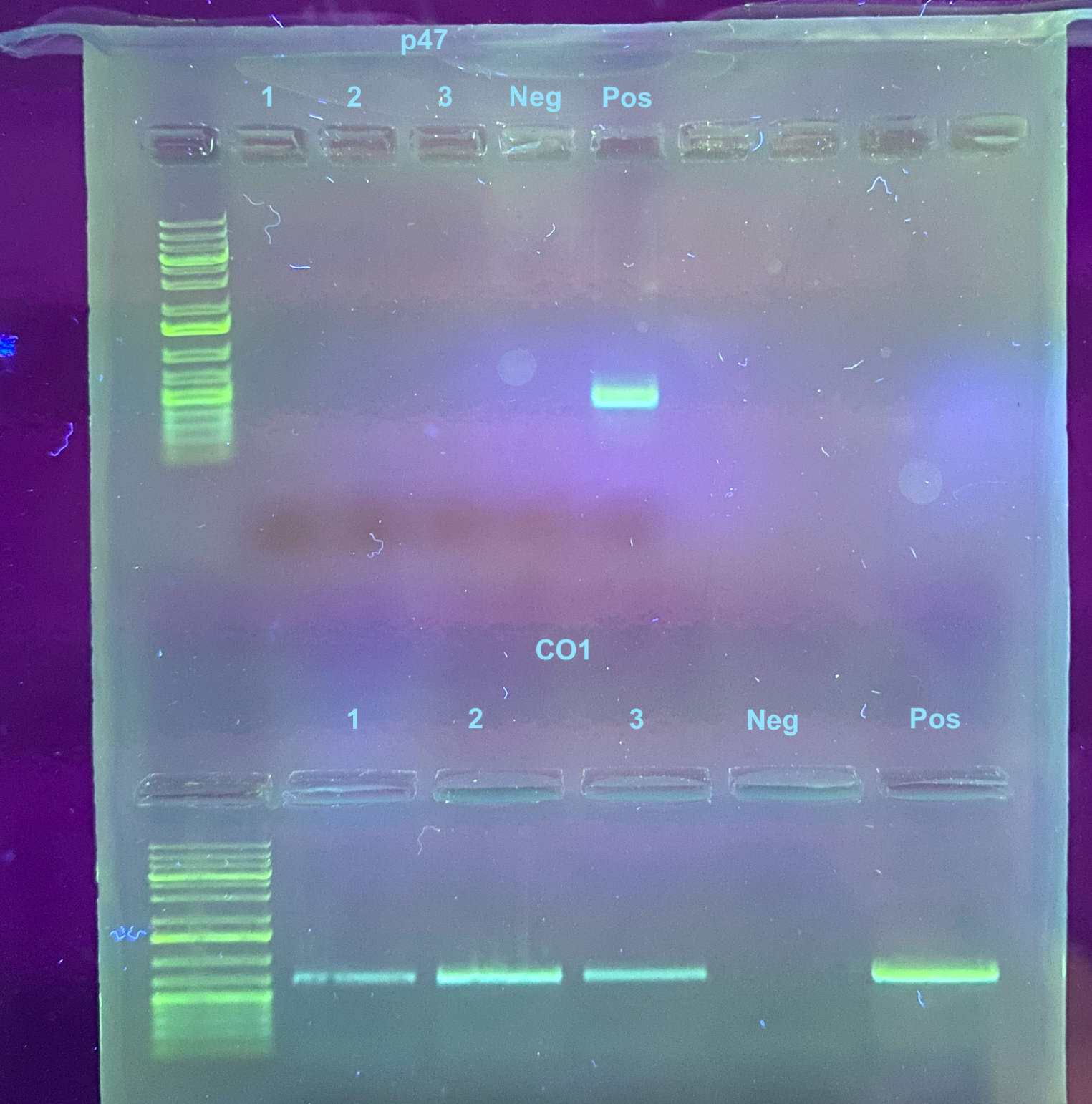
The cells are negative for virus based off of this PCR.