Testing 1ng, 0.1ng, and 0.02ng of DNA as input for qPCR
Previous qPCR showed that even for samples we thought had no infection/did not amplify and DiNV with traditional PCR, the Cq values for 115 were still mostly below 30. We would like to have virus negative samples have a high Cq value for virus primer if not 40.
It could be that the 1ng input I used was too much DNA and there could be spurious amplification. So I decided to try 1ng, 0.1ng, and 0.02ng input into the qPCR reaction.
I also tried this on a subset of samples, some with virus infection, and some of my stock innubila (not part of an infection) that had no amplification of DiNV with PCR.
Samples
| sample # | project | treatment | day frozen | p47 35 cycle result |
|---|---|---|---|---|
| 4 | stock innubila | NA | NA | none |
| 11 | stock innubila | NA | NA | none |
| 15 | stock innubila | NA | NA | none |
| 10 | infection | sterile poke | day 0 | strong band |
| 16 | infection | DiNV | day 0 | none |
| 37 | infection | sterile poke | day 3 | weak band |
| 42 | infection | DiNV | day 3 | strong band |
| 98 | infection | DiNV | day 5 | strong band |
| 143 | infection | sterile pole | day 10 | strong band |
| 150 | infection | DiNV | day 10 | strong band |
The 3 stock innubila samples needed to be Qubited and diluted to 1ng/ul. The other samples had already been diluted to 1ng/ul. All samples are thawed on ice and kept on ice. Samples are vortexed and spun down before use.
Qubit protocol is here and was followed exactly.
| sample | qubit | ul DNA needed for 1ng/ul | ul DNA hydration solution |
|---|---|---|---|
| 4 | 9.84 ng/ul | 3ul | 36.5ul |
| 11 | 30 ng/ul | 3ul | 87ul |
| 15 | 24.3 ng/ul | 3ul | 70ul |
For dilutions to 1:10 from 1ng/ul, so 0.1ng/ul
- All DNA thawed and kept on ice, as well as all dilutions
- Vortexed and spun down samples
- Made a set of strip tubes with 36ul of DNA hydration solution
- Added 4ul of appropriate DNA (at 1ng/ul) into it’s dilution tube
- Vortexed and spun down tubes and kept on ice
For dilutions 1:50 from 1ng/ul, so 0.02ng/ul
- All DNA thawed and kept on ice, as well as all dilutions
- Vortexed and spun down samples
- Made a set of strip tubes with 147ul of DNA hydration solution
- Added 3ul of appropriate DNA (at 1ng/ul) into it’s dilution tube
- Vortexed and spun down tubes and kept on ice
Here is the qPCR layout:
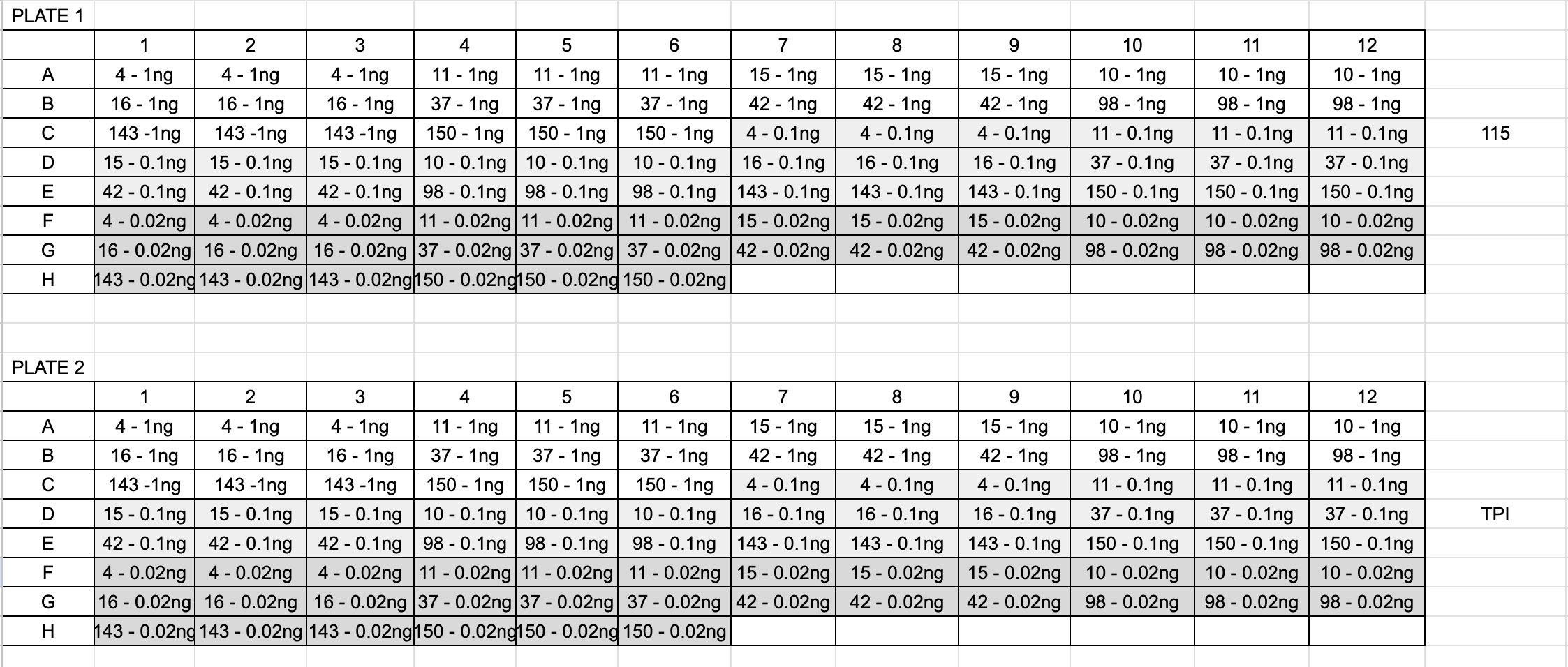
- Samples and reagents were thawed on ice, vortexed, and spun down before use
- There were 90 individual wells for each primer, 4 were added on for pipetting error
- 115 master mix:
- 5ul Sso supermix * 94 = 470ul
- 0.5ul 115 F primer * 94 = 47ul
- 0.5ul 115 R primer * 94 = 47ul
- 3ul nuclease free water * 94 = 282ul
- The master mix was made on ice, vortexed, and spun down
- TPI master mix:
- 5ul Sso supermix * 94 = 470ul
- 0.5ul TPI F primer * 94 = 47ul
- 0.5ul TPI R primer * 94 = 47ul
- 3ul nuclease free water * 94 = 282ul
- The master mix was made on ice, vortexed, and spun down
- I used the qPCR specific plates and seals
- 9ul of the appropriate master mix was added to the planned well (see layout above)
- 1ul of DNA was added to the planned well (see table above)
- Each well was pipette mixed with 5ul using a multichannel
- The plate was sealed and centrifuged for 2 minutes at 3,000g
- The plate was put in the qPCR machine:
- Used program CFX manager
- Selected user-defined protocol, existing protocol, and KMM folder
- Selected the p47 program
- Used the machine buttons to open and close the lid
- Started the program
- Data from the qPCR machine can be found here
- And analysis of the Cq results can be found here
Here is some diagnostic info (values are rounded), mean Cq is for 115 (virus primer). Note that PCR band results were from un-diluted/non-standardized DNA.
| sample # | project | treatment | day frozen | p47 35 cycle result | p47 30 cycle result | 1ng mean Cq | 0.1ng mean Cq | 0.02ng mean Cq | 1ng 2^delta Cq | 0.1ng 2^ delta Cq | 0.02ng 2^ delta Cq |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | stock innubila | NA | NA | none | NA | 32.6 | 33.3 | 33.9 | 0.0014 | 0.0062 | 0.021 |
| 11 | stock innubila | NA | NA | none | NA | 30.0 | 30.8 | 32.4 | 0.018 | 0.066 | 0.107 |
| 15 | stock innubila | NA | NA | none | NA | 31.6 | 31.5 | 33.5 | 0.003 | 0.034 | 0.027 |
| 10 | infection | sterile poke | day 0 | strong band | weak band | 27.8 | 28.9 | 30.9 | 0.028 | 0.109 | 0.103 |
| 16 | infection | DiNV | day 0 | none | none | 29.5 | 30.7 | 34.5 | 0.013 | 0.044 | 0.074 |
| 37 | infection | sterile poke | day 3 | weak band | none | 30.1 | 31.3 | 32.6 | 0.0096 | 0.0209 | 0.048 |
| 42 | infection | DiNV | day 3 | strong band | strong band | 21.9 | 25.0 | 27.4 | 4.2 | 1.8 | 2 |
| 98 | infection | DiNV | day 5 | strong band | medium band | 26.7 | 29.6 | 31.8 | 0.102 | 0.084 | 0.086 |
| 143 | infection | sterile poke | day 10 | strong band | medium band | 31 | 32.8 | 34.7 | 0.0062 | 0.016 | 0.02 |
| 150 | infection | DiNV | day 10 | strong band | strong band | 17.2 | 20.3 | 22.5 | 315 | 148 | 129 |