Re-doing a few PCRs for Freezing Experiment 1 to send some samples off for Sanger Sequencing
We don’t know if some of the positive virus samples from this project have junk DNA amplifying, or if it really is p47. So I re-did the PCRs in two ways, I either used the DNA un-diluted and ran it at 35 cycles for p47 (gives me the most bands), or I diluted each sample to 10ng/ul and ran it on 30 cycles.
A subset of these samples ended up getting used (see below):
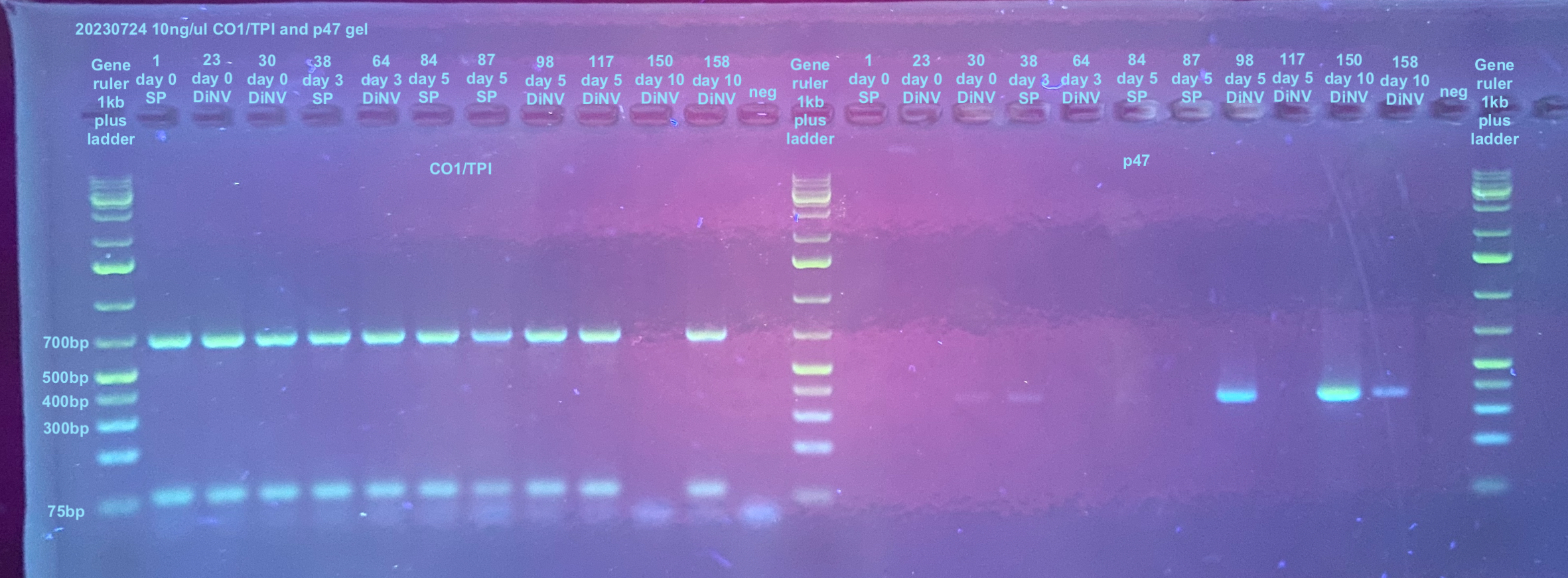
| sample_ID | treatment | p47_result | p47_30_cycle_result | qubit | total ng in 2ul | vol needed for 10ng/ul |
|---|---|---|---|---|---|---|
| 1 | sterile poke | yes | no | 31.3 | 187.8 | 18.78 |
| 23 | 16Cq DiNV | yes | no | 30.6 | 183.6 | 18.36 |
| 30 | 16Cq DiNV | yes | yes | 44.7 | 268.2 | 26.82 |
| 38 | sterile poke | yes | yes | 55.5 | 333 | 33.3 |
| 64 | 16Cq DiNV | yes | no | 67.6 | 405.6 | 40.56 |
| 84 | sterile poke | yes | yes | 38.4 | 230.4 | 23.04 |
| 87 | sterile poke | maybe | no | 102 | 612 | 61.2 |
| 98 | 16Cq DiNV | yes | yes | 43.8 | 262.8 | 26.28 |
| 117 | 16Cq DiNV | maybe | no | 51.3 | 307.8 | 30.78 |
| 150 | 16Cq DiNV | yes | yes | 50 | 300 | 30 |
| 158 | 16Cq DiNV | yes | yes | 37 | 222 | 22.2 |
20230724 10ng 30 cycle PCR
I re-checked these with CO1 and TPI because I had diluted them.
Filter tips were used in the making of the PCRs but not in the running of the gels.
PCRs on CO1, TPI, and p47 were done accordingly to the general PCR protocol with the primers, and their information can be found here. The p47 samples were amplified in 30 cycles.
A 2% gel was run for 45 minutes at 90V to analyze the PCRs. (see agarose gel protocol)
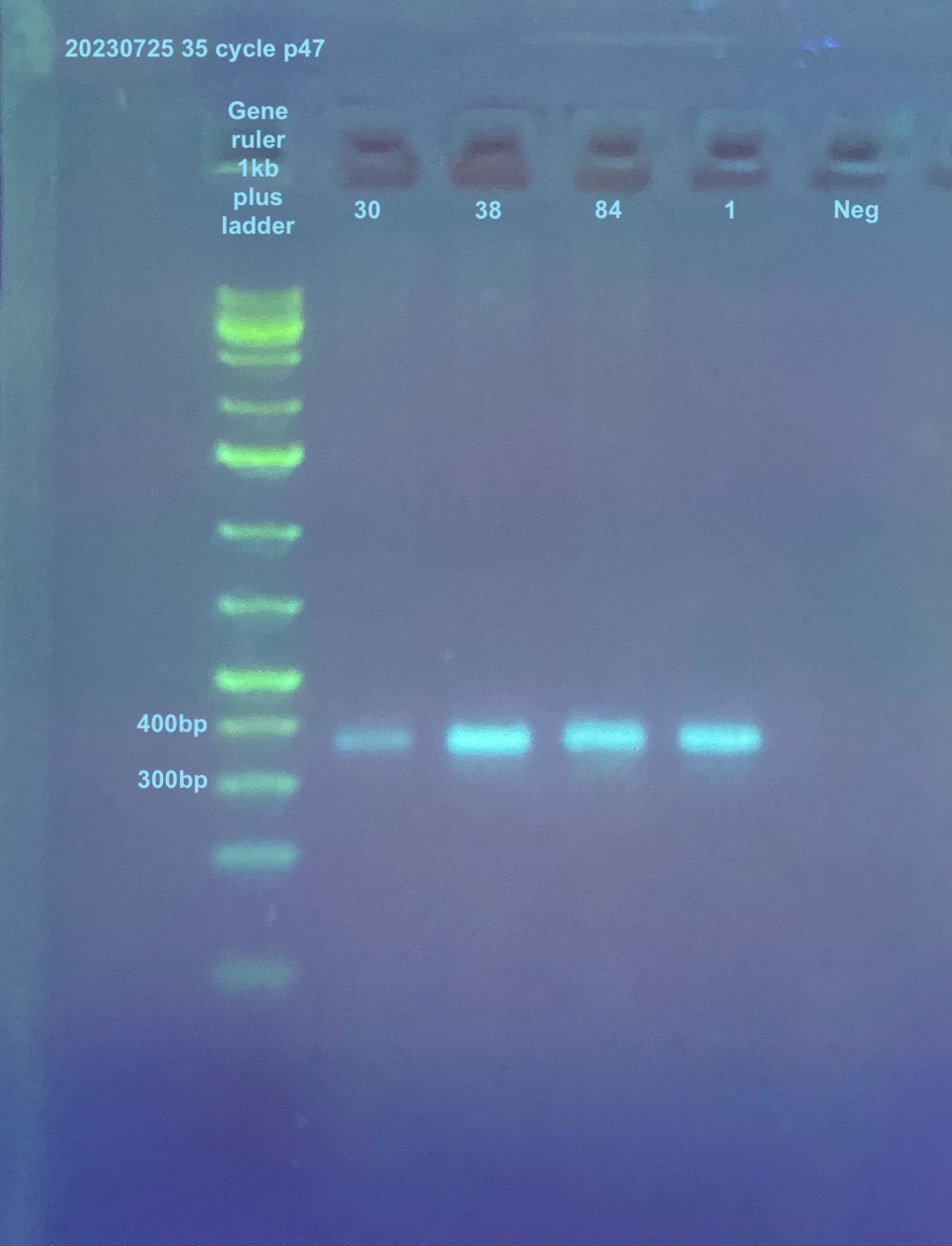
20230725 more p47 PCR
Because the 10ng and 30 cycle PCR had some of the samples not show bands where there had been using un-diluted samples, I decided to re-do the un-diluted 35 cycle p47 PCR on samples 1, 30, 38, and 84. Then I can compare those bands to the ones in the above PCR.
Again filter tips were used in the making of the PCRs but not in the running of the gels. I did not do CO1 or TPI for these samples because they had been done twice now.
PCRs on p47 was done accordingly to the general PCR protocol with the primers, and their information can be found here. The p47 samples were amplified in 35 cycles.
A 2% gel was run for 45 minutes at 90V to analyze the PCRs. (see agarose gel protocol)
20230726 Preparing Samples for Sequencing
These were the samples chosen for sequencing:
| tube | sample ID | type | band | treatment |
|---|---|---|---|---|
| 1 | 1 | 10ng 30 cycle | no | SP |
| 2 | 1 | 35 cycle non-diluted | strong | SP |
| 3 | 30 | 10ng 30 cycle | weak | DiNV |
| 4 | 30 | 35 cycle non-diluted | medium | DiNV |
| 5 | 38 | 10ng 30 cycle | weak | SP |
| 6 | 38 | 35 cycle non-diluted | strong | SP |
| 7 | 84 | 10ng 30 cycle | ? | SP |
| 8 | 84 | 35 cycle non-diluted | strong | SP |
| 9 | 98 | 10ng 30 cycle | strong | DiNV |
| 10 | 150 | 10ng 30 cycle | strong | DiNV |
| 11 | 158 | 10ng 30 cycle | weak | DiNV |
All PCR products pere prepared accordingly to the prep samples for sanger sequencing protocol