Testing the Ability to Do Multiplex PCRs of CO1 and TPI, and 115 and p47
I want to test if I can run CO1 and TPI primers in on PCR reaction, and 115 and p47 primers in also the same PCR reaction. This will allow me to test 4 primers on my DNA extractions, while only doing 2 actual reactions per sample.
For all of these tests I will be using samples 51, 38, 11, and 9 from the 20220915 innubila DiNV infections, where 51 and 38 are virus positive, and 11 and 9 are virus negative.
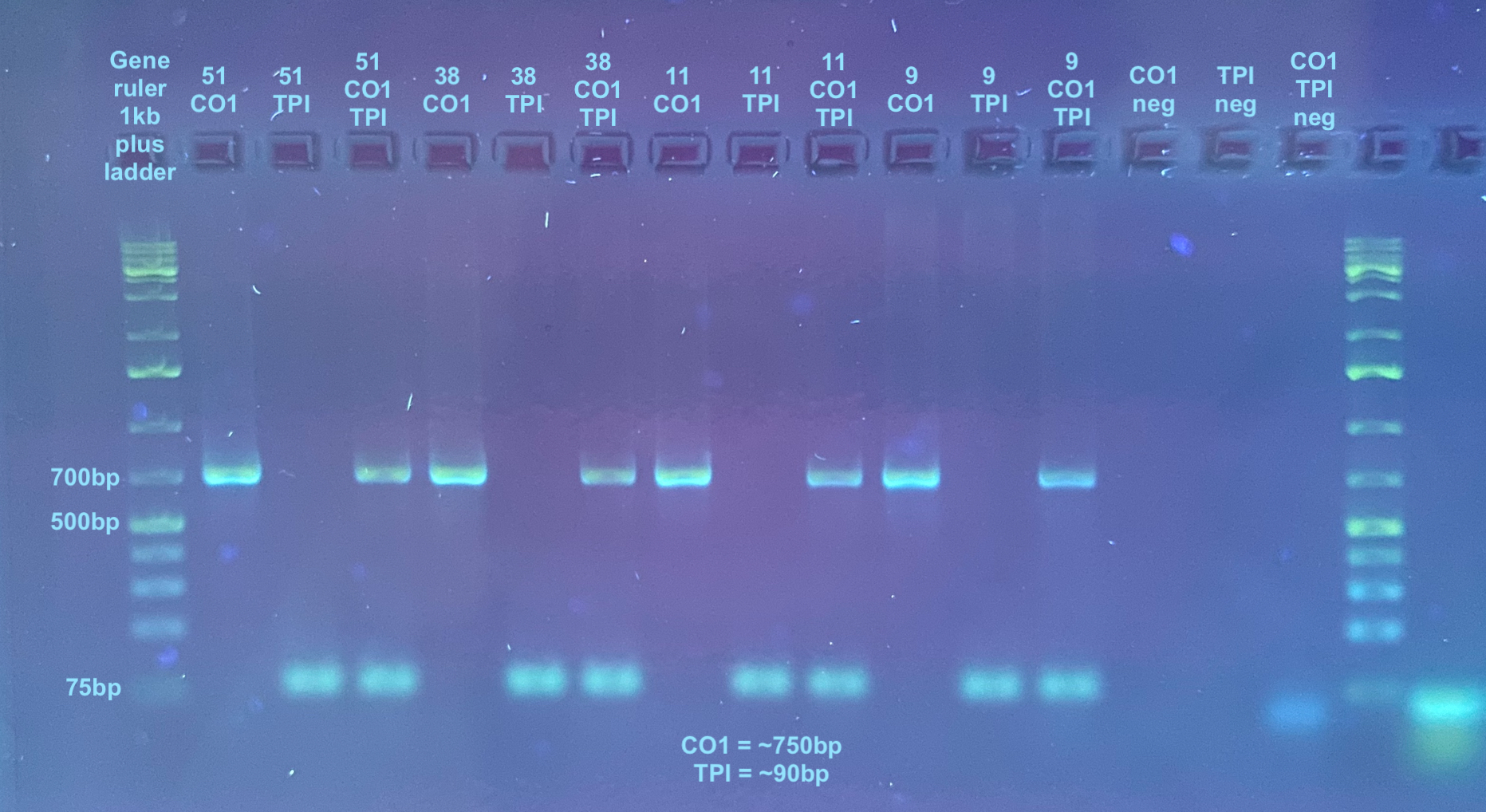
CO1/TPI
- CO1 is about 750bp and TPI is about 90bp, so they can both be resolved on the same gel
- The primers both have their lowest TMs at 59, so the same annealing temp should be able to be used
- I will use the CO1 program to test these out:
- 95 degrees C 2 min
- 95 degrees C 20 sec
- 52 degrees C 20 sec
- 72 degrees C 1 min 30 sec
- 72 degrees C 5 minutes
- 12 degree C hold
- bold lines are cycled 34 times
- Each sample was tested with CO1 primers only, TPI primers only, and both primers, as was a negative control
- All samples, primers, and reagents were thawed on ice, vortexed, and spund down before use
- Made 3 master mixes:
- CO1 master mix:
- 27ul GoTaq
- 19.25ul molecular grade water
- 1.37ul CO1 F primer
- 1.37ul CO1 R primer
- TPI master mix:
- 27ul GoTaq
- 19.25ul molecular grade water
- 1.37ul TPI F primer
- 1.37ul TPI R primer
- CO1 + TPI master mix:
- 27ul GoTaq
- 16.5ul molecular grade water
- 1.37ul CO1 F primer
- 1.37ul CO1 R primer
- 1.37ul TPI F primer
- 1.37ul TPI R primer
- All master mixes were vortexed and spun down and kept on ice during use
- 9ul of the appropriate master mix was added to each tube:
| tube number | sample | primer |
|---|---|---|
| 1 | 51 | CO1 only |
| 2 | 51 | TPI only |
| 3 | 51 | TPI and CO1 |
| 4 | 38 | CO1 only |
| 5 | 38 | TPI only |
| 6 | 38 | TPI and CO1 |
| 7 | 11 | CO1 only |
| 8 | 11 | TPI only |
| 9 | 11 | TPI and CO1 |
| 10 | 9 | CO1 only |
| 11 | 9 | TPI only |
| 12 | 9 | TPI and CO1 |
| 13 | neg | CO1 only |
| 14 | neg | TPI only |
| 15 | neg | TPI and CO1 |
- Added 1ul of the appropriate DNA sample or molecular grade water for the controls
- Vortexed and spun down the tubes
- Placed the tubes in the CO1 program (listed above)
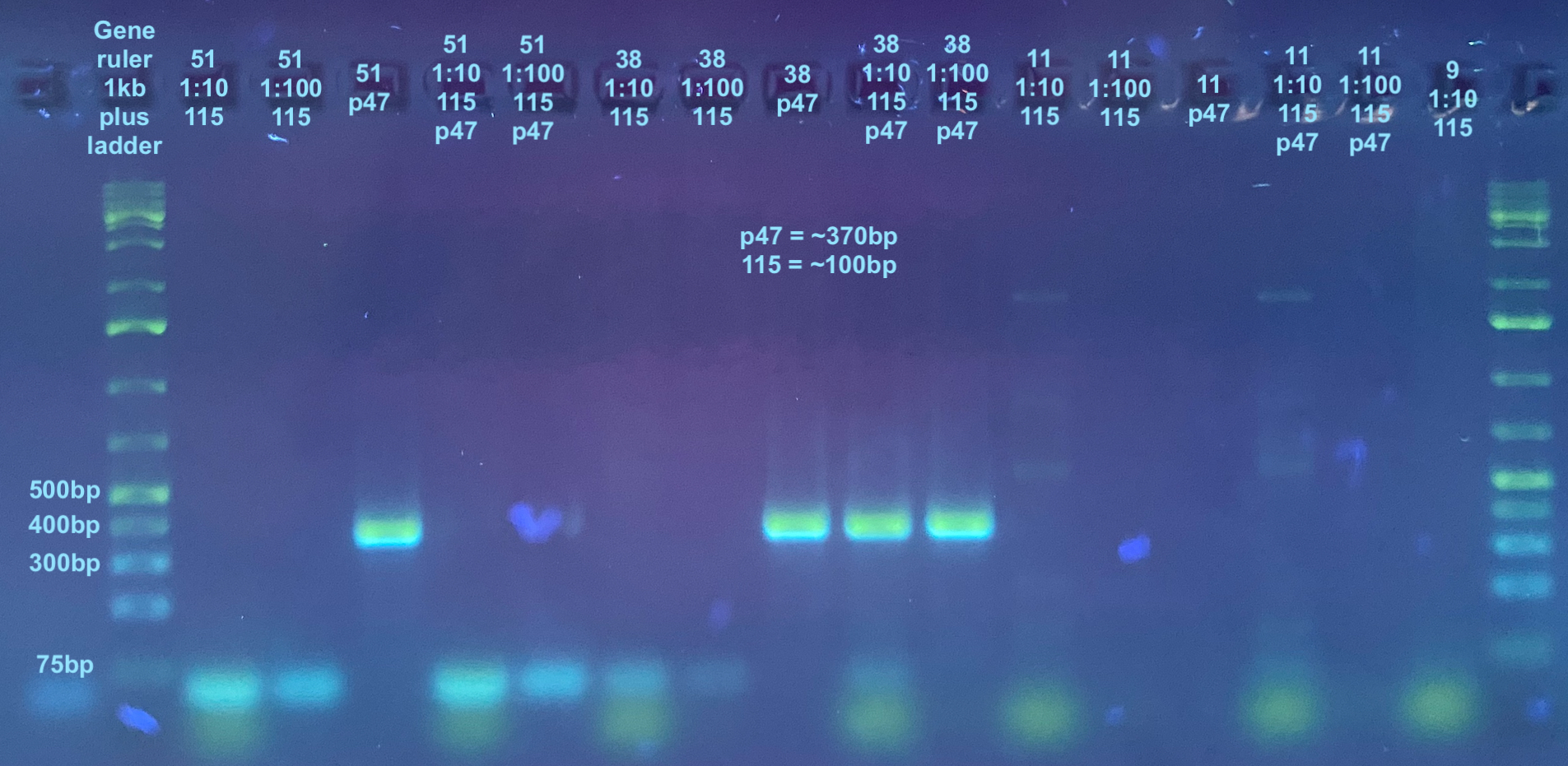
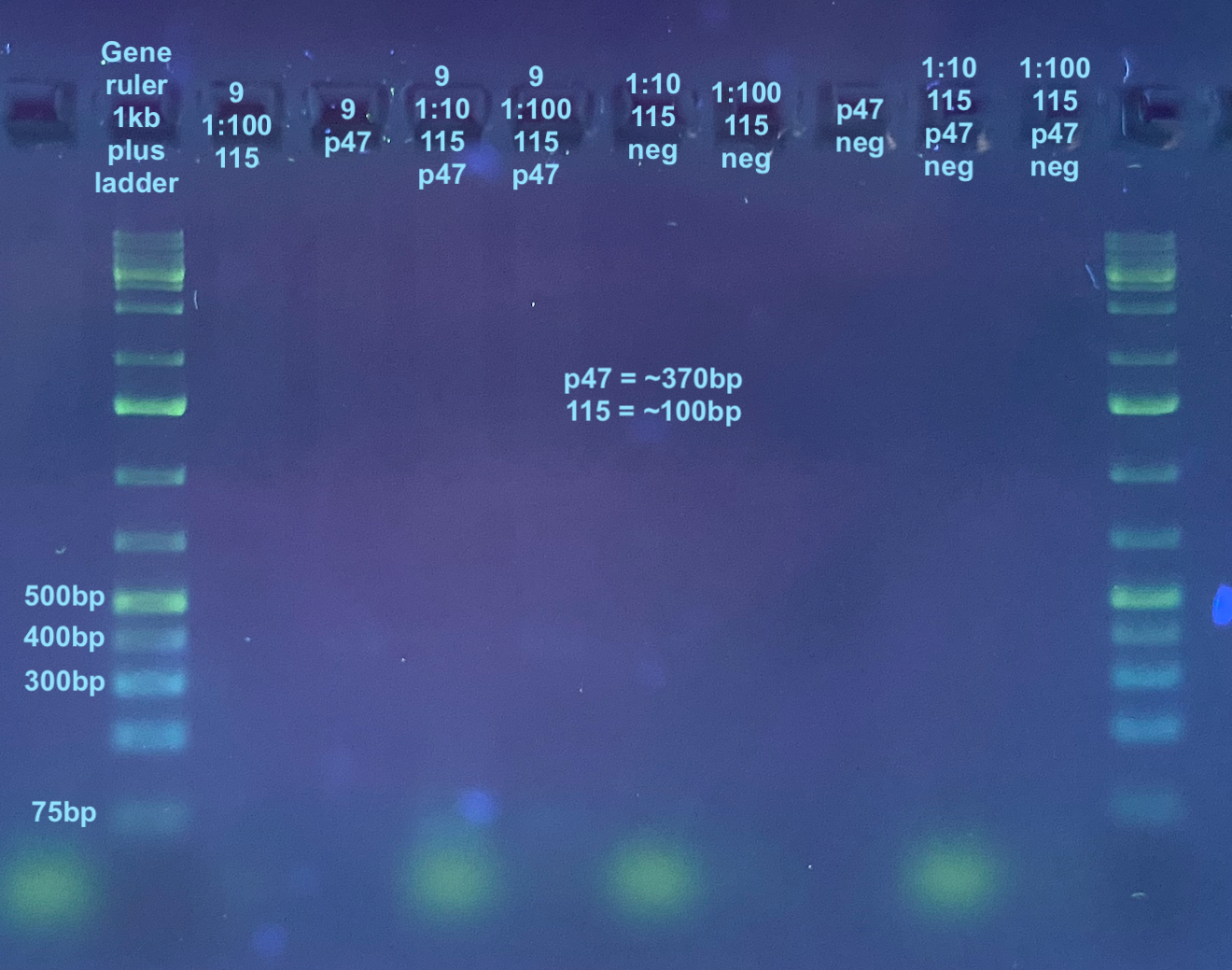
115 and p47
- 115 is about 100bp and p47 is about 370bp so they should be able to be resolved on a gel togther
- Both PCR programs use a 55C annealing temp, I decided to go with the 115 program for this trial:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 55 degrees C 30 sec
- 72 degrees C 30 sec
- 72 degrees C 5 minutes
- 12 degree C hold
- bold lines are cycled 34 times
- For this I used Kent’s 115 stock primers that may need to be diluted 1:100 instead of 1:10 to get 10uM concentration. I decided to test both dilutions of the primers
- Each sample was tested with p47 primers only, 115 primers only (both 1:10 and 1:100), and both combo sets of primers, as was a negative control
- All samples, primers, and reagents were thawed on ice, vortexed, and spund down before use
- Made 5 master mixes:
- p47 master mix:
- 27ul GoTaq
- 19.25ul molecular grade water
- 1.37ul p47 F primer
- 1.37ul p47 R primer
- 115 1:10 master mix:
- 27ul GoTaq
- 19.25ul molecular grade water
- 1.37ul 115 1:10 F primer
- 1.37ul 115 1:10 R primer
- 115 1:100 master mix:
- 27ul GoTaq
- 19.25ul molecular grade water
- 1.37ul 115 1:100 F primer
- 1.37ul 115 1:100 R primer
- p47 + 115 1:10 master mix:
- 27ul GoTaq
- 16.5ul molecular grade water
- 1.37ul p47 F primer
- 1.37ul p47 R primer
- 1.37ul 115 1:10 F primer
- 1.37ul 115 1:10 R primer
- p47 + 115 1:100 master mix:
- 27ul GoTaq
- 16.5ul molecular grade water
- 1.37ul p47 F primer
- 1.37ul p47 R primer
- 1.37ul 115 1:100 F primer
- 1.37ul 115 1:100 R primer
- All master mixes were vortexed and spun down and kept on ice during use
- 9ul of the appropriate master mix was added to each tube:
| tube number | sample | primer |
|---|---|---|
| 16 | 51 | 1:10 115 only |
| 17 | 51 | 1:100 115 only |
| 18 | 51 | p47 only |
| 19 | 51 | p47 and 1:10 115 |
| 20 | 51 | p47 and 1:100 115 |
| 21 | 38 | 1:10 115 only |
| 22 | 38 | 1:100 115 only |
| 23 | 38 | p47 only |
| 24 | 38 | p47 and 1:10 115 |
| 25 | 38 | p47 and 1:100 115 |
| 26 | 11 | 1:10 115 only |
| 27 | 11 | 1:100 115 only |
| 28 | 11 | p47 only |
| 29 | 11 | p47 and 1:10 115 |
| 30 | 11 | p47 and 1:100 115 |
| 31 | 9 | 1:10 115 only |
| 32 | 9 | 1:100 115 only |
| 33 | 9 | p47 only |
| 34 | 9 | p47 and 1:10 115 |
| 35 | 9 | p47 and 1:100 115 |
| 36 | neg | 1:10 115 only |
| 37 | neg | 1:100 115 only |
| 38 | neg | p47 only |
| 39 | neg | p47 and 1:10 115 |
| 40 | neg | p47 and 1:100 115 |
- Added 1ul of the appropriate DNA sample or molecular grade water for the controls
- Vortexed and spun down the tubes
- Placed the tubes in the 115 program (listed above)
Gel
- Made a large 2% gel with:
- 3.3g of agarose
- 165mL of 1X TAE
- 2ul midori stain
- Gel ran for 50min at 90V
CO1 and TPI
p47 and 115