Planning for Cloning DiNV With pSPIN By Designing BAC and pSPIN
Begin with checking pSPIN/pSL1142 for the restriction sites where the donor DNA/BAC can be inserted
- This is all done in Geneious where I have a good annotated file of the plasmid
- The left side of the insert site has a Xho1 restriction site
- The left side of the insert site has a Pst1 restriction site
- We can’t go any further into the plasmid because we need to keep the VchINT sites for the integration to happen (sites needed for transposition)
Checking our two BACs for those restriction sites
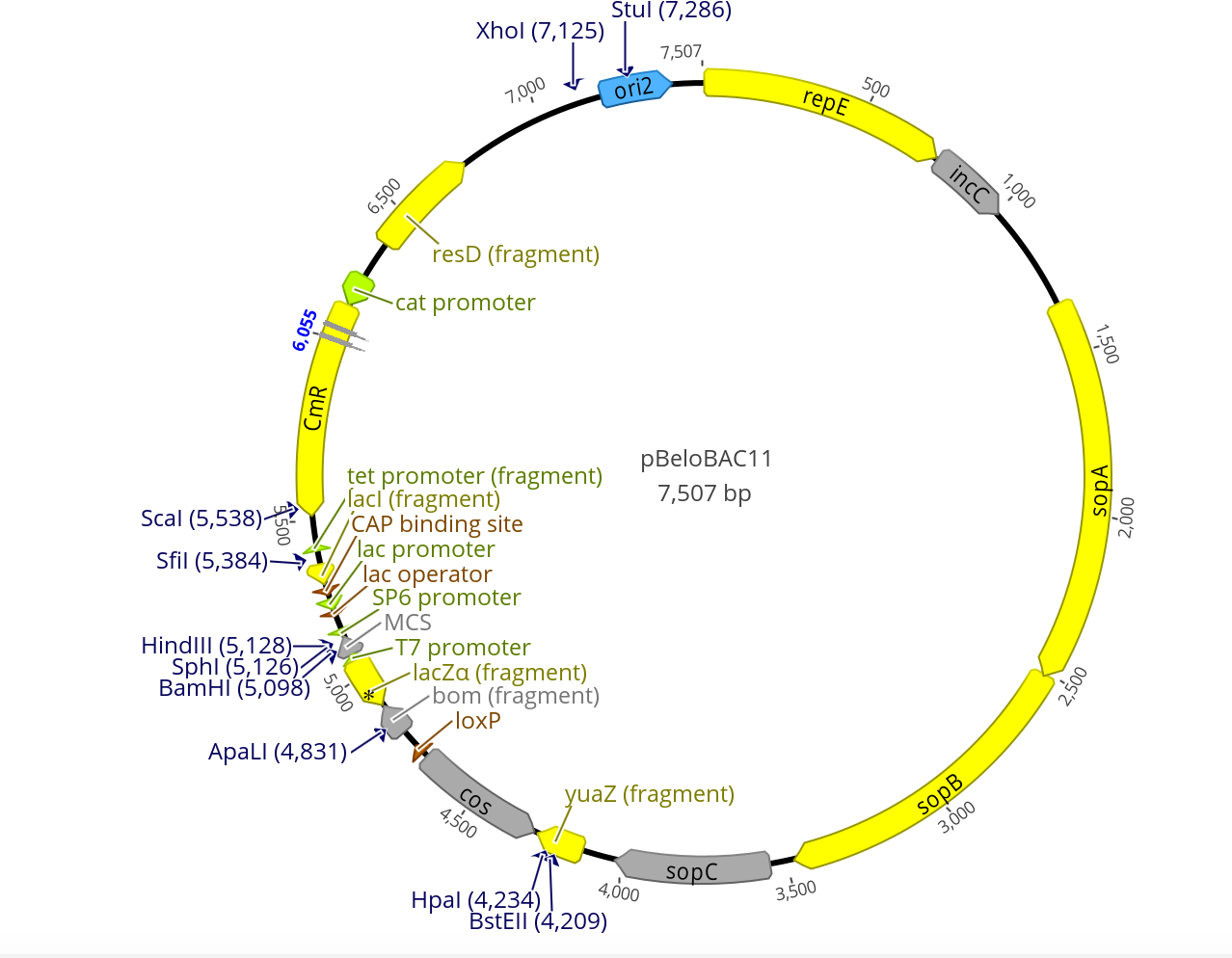
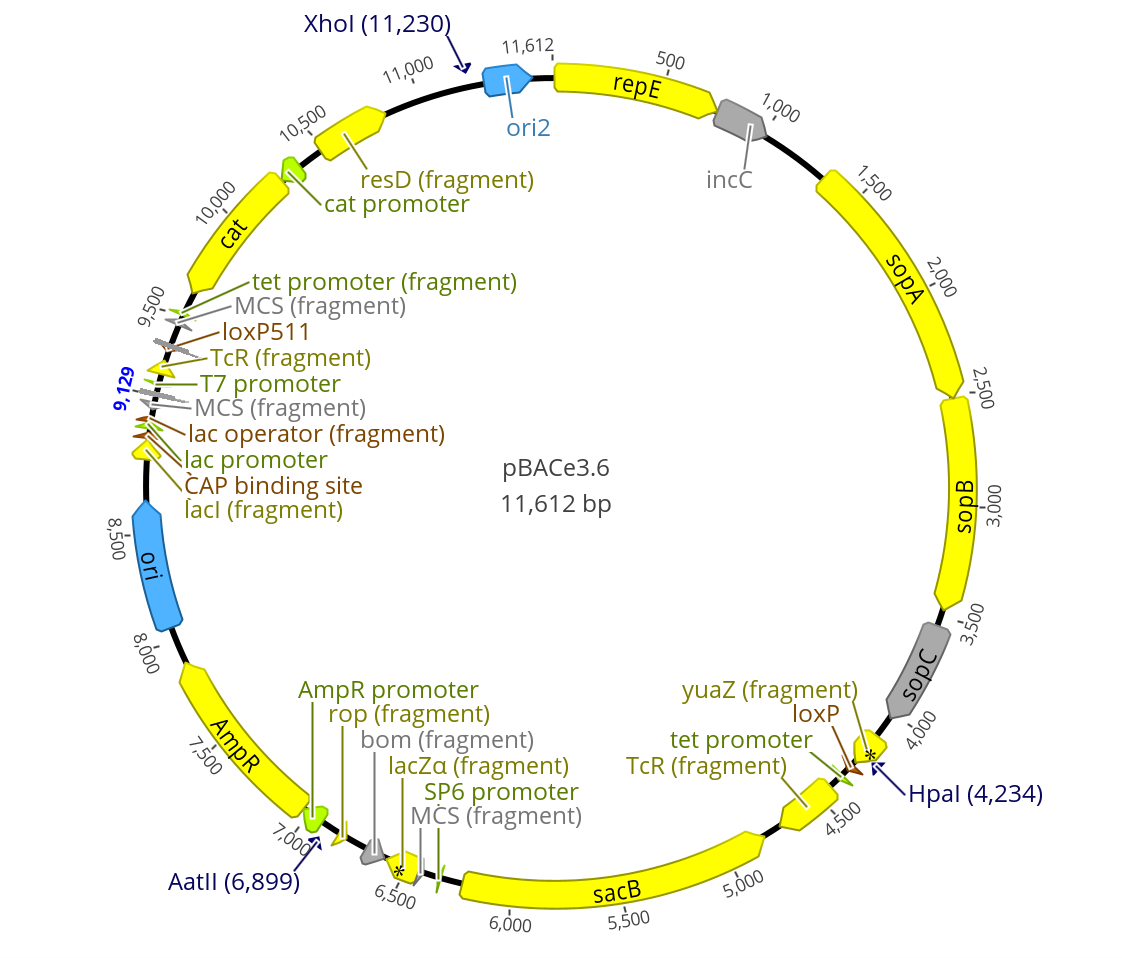
- Both pBeloBAC11 and pBACe3.6 have a Xho1 restriction site near their ORI2
- Neither have a Pst1 restriction site
- I’m not sure at this point if we need both restriction sites or not to clone in the BAC
- In general, pBACe3.6 has less restriction sites than pBeloBAC11
Comparing BACs
- pBeloBAC11 has Chloramphenicol resistance
- pBACe3.6 has both Chloramphenicol and Ampicillin resistance
- pBAC3e3.6 is 11,612bp
- pBeloBAC11 is 7,507bp
- Maybe it is better to start with a smaller BAC because we will have to add in things to it? if pAc5 promoter ends up working, that is ~2,500bp not including the fluorescence gene
What are things needed in the BAC?
- Resistance/selectable marker
- We have this in either BAC
- Fluorescence marker
- Low priority if next weeks transfection results are inconclusive/negative
- Recombinase sites
- This I am unsure how to design/place
- Site for cloning into pSPIN
Where to put in the recombinase and potential Fluorescence marker in the BAC
- If we are stuck with the Xhol1 as the cloning site, then I’m not sure how we can get the recombinase sites and marker in
- Restriction sites highlighted pBeloBAC11:
- Restriction sites highlighted pBACe3.6:
- However, maybe we can have genewix clone into the BAC the marker, recombinase sites, as well as the sites for cloning into pSPIN, so it all fits?
What does the pSPIN need?
- Selectable marker
- Has Kanamycin resistance
- Designed BAC inserted
- crRNA sequence inserted
Revisiting the crRNA sequences
Reminder that efficiency score comes from this website and can only take 20bp of sequence. Everything else comes from BLASTing the sequence to various species databases.
| # | sequence | Location | Location issue? | BLAST to e coli? | BLAST to 2nd place in DiNV | BLAST to D. virilis | BLAST to D. innubila | BLAST to D. falleni | Efficiency score of first 20bp | Efficiency score of last 20bp | Repetitive in 10bp on 3’ end? |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CCCTGAATATTTTCTAGCCATCGCGCACCCAT | 76903 | 160bp downstream of a gene | N | N | N | N | N | 3.74 | 7.649 | N |
| 1 | TCGCTGCCATTATACTCACAATCACATCGAGT | 78530 | In a gene | N | N | N | N | N | 3.86 | 3.27 | N |
| 2 | CCCCCCTCGAATGTTACTTTTGTACTATATAT | 78077 | N | N | N | N | N | N | 8.08 | 5.16 | Y? |
| 3 | AATAAAAATCATCGCACCCTCTCCCACACTGA | 77423 | N | N | N | N | N | N | 7.3 | 4.94 | N |
| 4 | CCTGAATATTTTCTAGCCATCGCGCACCCATC | 76904 | 160bp downstream of a gene | N | N | N | N | N | 6.14 | 6.14 | N |
| 5 | CTGAATATTTTCTAGCCATCGCGCACCCATCT | 76905 | 160bp downstream of a gene | N | N | N | N | N | 4.63 | 4.67 | N |
| 6 | TGAATATTTTCTAGCCATCGCGCACCCATCTC | 76906 | 160bp downstream of a gene | N | N | N | N | N | 3.99 | 4.21 | N |
| 7 | ATCGCGCACCCATCTCTAAATCGCGCAACGAC | 76922 | 180bp downstream of a gene | N | N | N | N | N | 5.78 | 7.13 | N |
| 8 | CATCTCTAAATCGCGCAACGACTTGATCAATG | 76932 | 190bp downstream of a gene | N | N | N | N | N | 4.91 | 6.32 | N |
| 9 | ATCTCTAAATCGCGCAACGACTTGATCAATGT | 76933 | 190bp downstream of a gene | N | N | N | N | N | 3.34 | 6.29 | N |
| 10 | CTCTCCCACACTGACAATACATCTATTTCCCC | 77405 | N | N | Y | N | N | N | 5.65 | 5.01 | Y |
| 11 | TCTCCCACACTGACAATACATCTATTTCCCCA | 77404 | N | N | Y | N | N | N | 6.04 | 6.97 | N |
| 12 | CACACTGACAATACATCTATTTCCCCAAGTAT | 77399 | N | N | Y | N | N | N | 5.04 | 3.99 | N |
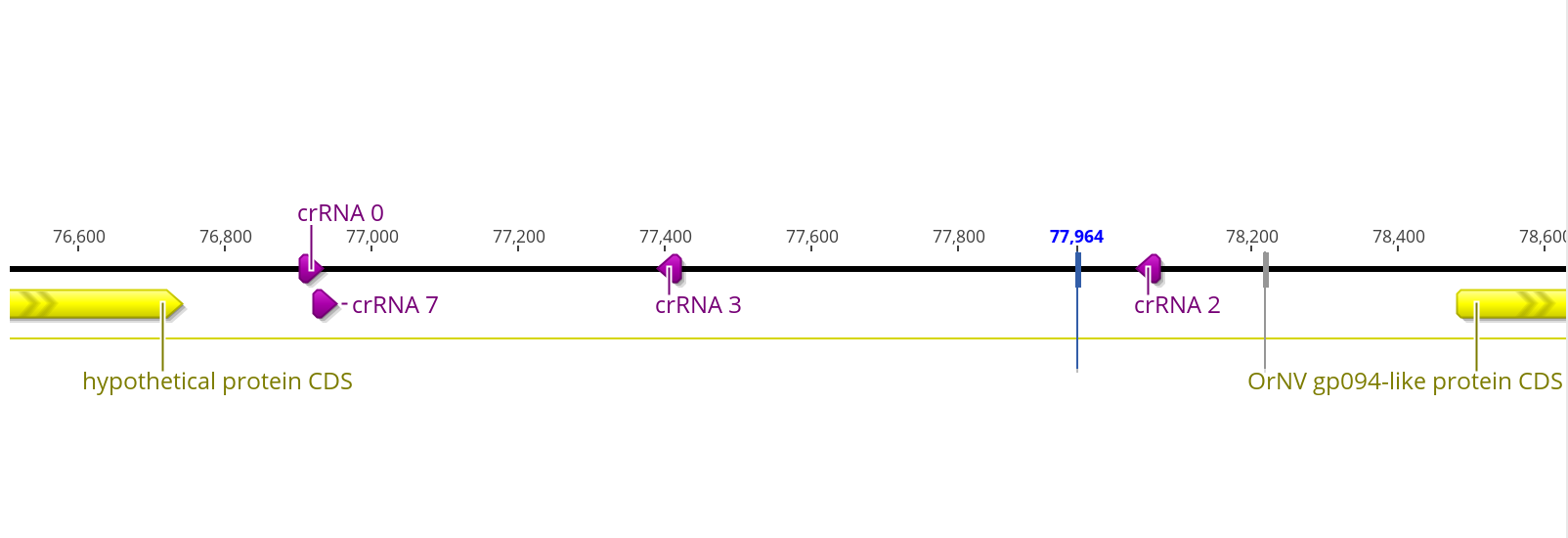
Based off of this, we are most interested in in # 0, 7, and maybe 2 and 3. Where are these crRNA sites in DiNV?
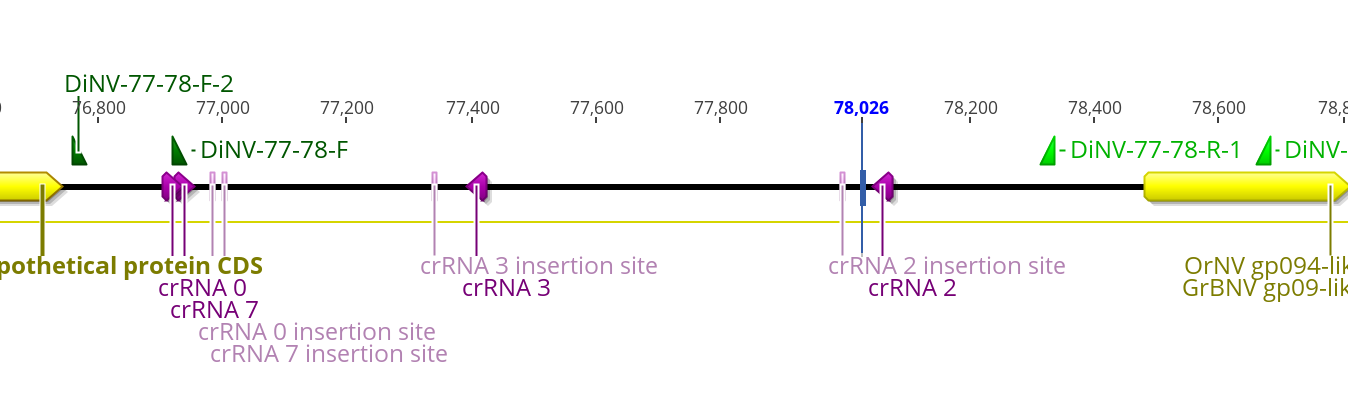
Ok and how do these crRNA sites line up to primers I already have used in this region of DiNV?
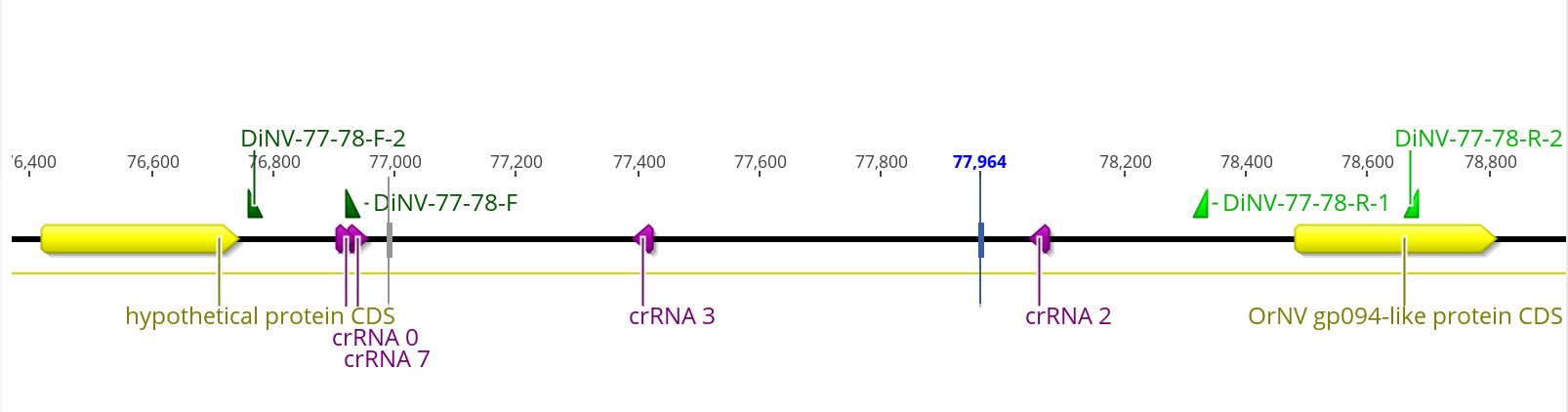
The primers I already have may work, it is hard to imagine what this area will look like once the BAC is inserted into it. The goal would be to have primers that start on the DiNV side, and go into the BAC.
Here I have where I think the actual insertion sites might be. I put them 49bp downstream of the crRNA sequence… I am not sure if that is doing the proper orientation.