Using D. melanogaster Constitutive Promoters to Search For Promoters for D. innubila
I need to figure out a promoter to use to get GFP or RFP to express in innubila cells/flies.
Finding D. melaongaster promoter sequences
- Through some Google searching and talking with Rob, some good constitutive melanogaster promoters are:
- Actin 5 C
- Ubiquitin
- Tubulin
- U6
- COPIA
- EF-1-alpha
- Finding the sequences for these was actually quite hard, simple Googling the sequences does not give you them!
- So I started looking in Addgene to see if there were any plasmids that have these promoters in them to express whatever’s in the plasmid
- This got me most of the promoter sequences:
- Actin 5C
- Ac5-STABLE2-RFP-NLS-CycB(1-266)_GFP-E2F1(1-230)_neo
- pAc5.1B-EGFP-DmNot3
- These two plasmids have almost the same size sequence, there’s ~3bp difference. I ended up using the longer sequence
- This promoters is 2,516 bases long, which is really long
- Sequence link
- COPIA
- Hy_pMT Laccase2 MCS-WT Split GFP
- I only found one plasmid with this sequence
- This promoter is 280 bases long
- Sequence link
- U6
- pCFD3.1-w-dU6:3gRNA
- There were multiple plasmids that had this promoter
- There are multiple U6 promoters, but I found info on this website that says “U6:3, the strongest U6 promoter in Drosophila”
- This promoter is 421 bases long
- Sequence link
- Ubiquitin/poly-Ubiquitin promoter
- This one was pretty hard to find on Addgene, if you just search for “ubiqutin promoter” and specify d.mel expression, none of the images of the plasmids have anything in them that look like a promoter
- For example, if you look in the Addgene image for this plasmid pDmPolyUbiq, it doesn’t look like it has the promoter sequence annotated. If it’s not mapped out on the plasmid picture/genebank file then I can’t extract it
- But! If I do download the genbank file then the promoter region is annotated and I can extract it
- This promoter is 2,008 bases long
- Sequence link
- Tubulin
- pDmAlfaTub84b, this primer is like the one above where it’s image doesn’t show the promoter sequence, but the genebank file does
- This is the only plasmid I found with a good file for the promoter
- This promoter is 2,399 bases long
- Sequence link
- Ef-1-alpha
- I was not able to find this promoter in an Addgene plasmid. I started Googling again, and I found this paper that gives primers for amplifying the whole promoter sequence to clone it into a plasmid
- I used the primer sequences to BLAST to the melanogaster genome to get the whole sequence
- Based on the BLAST, the promoter starts at 11891257 on the 2R chromosome and ends at 11893572
- I went and looked on FlyBase and this does match where the EF-1-alpha gene is, but interestingly it completely overlaps with the gene, it’s basically the first ~2000 bases of the gene
- I wasn’t sure if that would mean it’s not actually the promoter sequence, but this is the only lead I could find
- I downloaded the gene sequence from FlyBase and separate out the putative promoter sequence. The gene sequence goes from 11895135 to 11895762
- Based off of the BLAST homology, the putative promoter starts 122 bases into the gene sequence, and ends 2437 bases in
- I used python to convert it all to uppercase, then remove the new lines. Then I separated out the actual promoter sequence
import os os.chdir("/Users/maggieschedl/Desktop/") alpha = open("dmel-E1-alpha.txt", ) alpha_contents = alpha.read() #print(alpha_contents) upper = alpha_contents.upper() # make uppercase # print(upper) # check uppercase # remove new lines alpha_contig = upper.replace('\n', '') #print(alpha_contig) first122 = alpha_contig[0:123] #print(first122) promoter = alpha_contig[123:2438] print(promoter) - This promoter is 2,315 bases long
- Sequence link
Checking sequences by BLASTing to D. mel genome
- Not that I don’t trust the things on Addgene, I just want to make sure that the promoter sequences that I got are actually D. mel and not human or something else. There were multiple plasmids I looked at that said insect and human expression (not sure how that is possible)
- I already know that Ef-1-alpha is melanogaster because the genome is where I got the sequence from
- I used the RefSeq Genome database and the highly similar sequences option
- Actin 5C
- Had 2 matches on the X chromosome (non-overlapping)
- First is from base 11 to 601 of the promoter
- Bases 5898377 to 5898972 on the chromosome
- Second is from base 611 to 2516 of the promoter (the end)
- Bases 5899048 to 5900947 on the chromosome
- This is pretty good coverage of the promoter, and it has a good match to the chromosome, but there is the ~76bp gap in between those two matches, but that doesn’t seem to large
- Is this relevant/does it matter for promoting? Unsure
- I then looked at the region it BLASTd to on FlyBase and it is right before the Act5C gene and a few bases at the start of it
- A big portion of the “promoter” is in the lncRNA sequence upstream of Act5C gene
- COPIA
- This BLASTd to a lot of places, multiple chromosomes
- This is probably because it is a transposon promoter (?)
- There are a lot of 280/280 exact matches, so my guess is that it is in a lot of places in the genome, and the promoter is accurate
- U6
- Had 1 match on the 3R chromosome
- Every base is an exact match except for a string of a few As in the middle of the sequence
- Bases 24556795 to 24557215 on the chromosome
- This area is right up to the start of snRNA:U6:96Ac. There are other snRNA:U6s more upstream. This seems to be a correct promoter
- Ubiquitin/poly-ubiquitin
- Had 1 match to the 3L chromosome
- Very close to an exact match, there are a few bases that are off in the middle, and it is missing about 16 bases at the end, but it looks very good
- Bases 3903655 to 3901760 on the chromosome
- Ubiquitin goes in the “reverse direction”, but this region is in front of it, and overlapping with the ubiquitin gene quite a bit (~1,400 bases)
- It also overlaps with the Sc2 gene for about 150 bases
- This is similar to the EF-1-alpha “promoter” where is significantly overlaps with the gene
- Tubulin
- Had 1 match to the 3R chromosome
- Very close match, only ~3 bases don’t match
- Bases 7084342 to 7086740 on the chromosome
- This is before a tubulin gene but it has a lot of overlap with the gene before it
- Overlaps ~1,700 bases in the Tailor gene at the end of it, and about 150 bases of the alphaTub84b gene at the beginning
BLASTing promoter sequences to D. innubila genome
- Now I want to see how much homology there is with these sequences and the innubila genome. Some of these sequences are very long so I’m not sure what I am going to get
- Using RefSeq genome database to innubila genome, and using somewhat similar sequence algorithm
- Actin 5C
- There are 5 matches, one to the X chromosome, and the others are to 3 and 2. I am only going to look at the X chromosome because that is where it is on melanogaster
- The best match on the X chromosome is about 113 bases, and there are a couple of gaps in it. It says in the features section that this is near the Actin 5C gene, so at least this seems to be the correct area
- There is a smaller match near the Actin 5C gene that is 36 bases, the rest of the matches seem to be in different parts of the chromosome
- The main match is from 26272271 to 26272154 on the innubila X chromosome
- The smaller match is from 26273932 to 26273896 on the innubila X chromosome
- These are upstream of innubila LOC117794475 which BLASTs very closely to the dmel Acting 5C gene (as well as some other Actin genes)
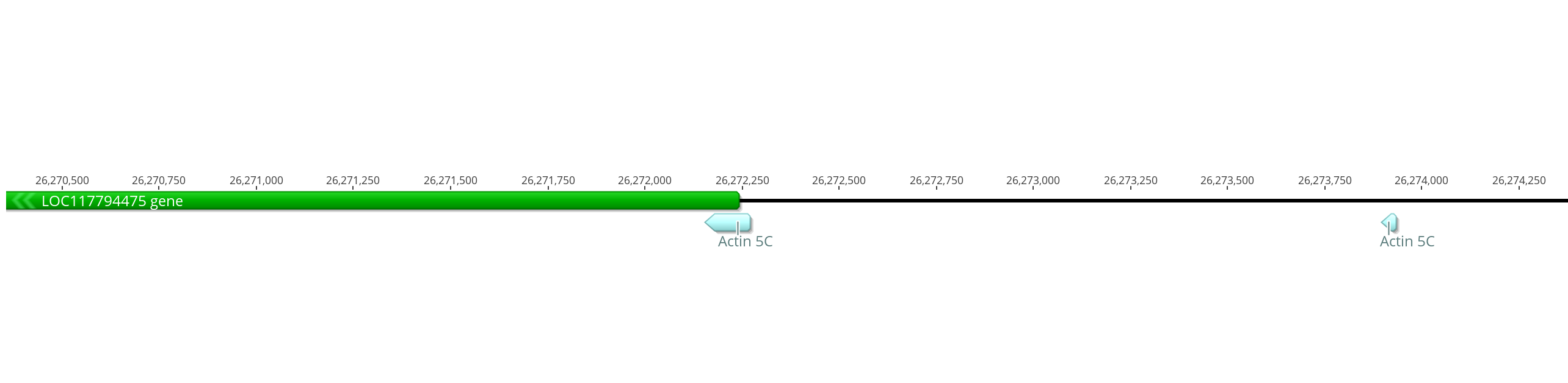
- Because there is just so little overlap from the dmel promoter to the innubila genome, I am just going to take this section of the innubila genome and assume this is the promoter. I tried to make it similar size and similar overlap into the actual gene as dmel has
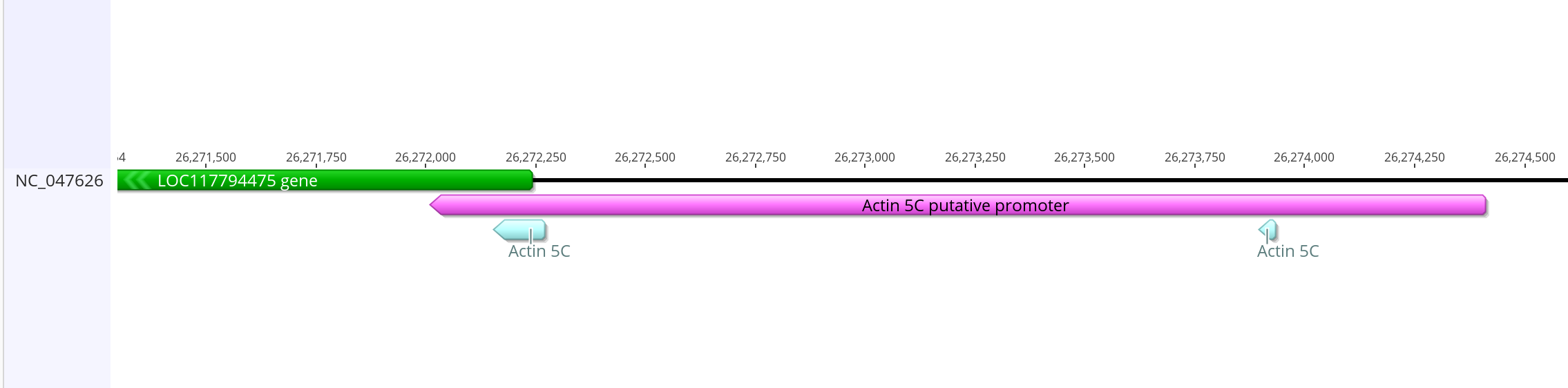
- This is how it compares with the mRNA from the gene
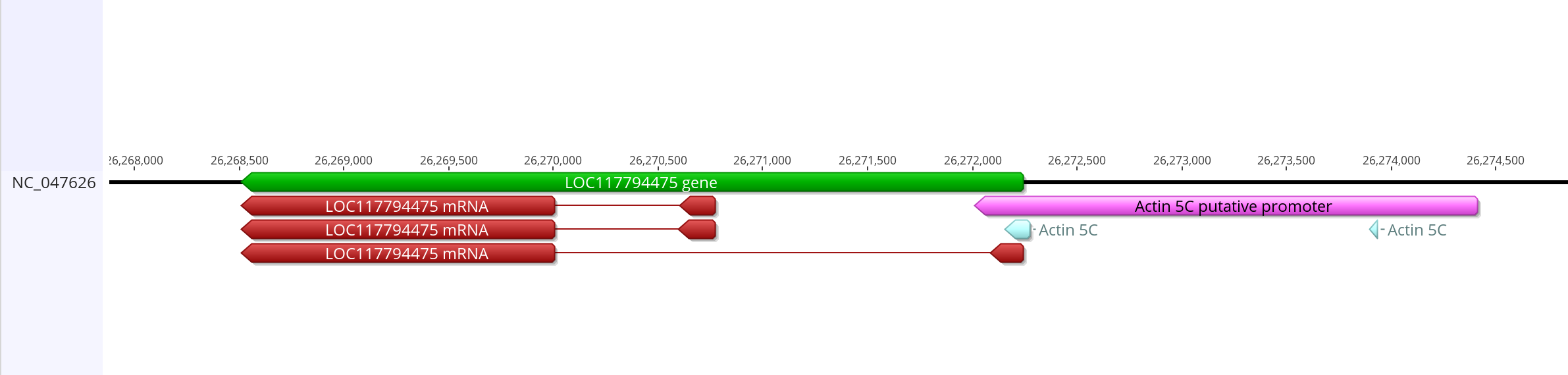
- Sequnece link
- This putative promoter is 2,401 bases
- COPIA
- There are matches to all chromosomes here, this might be a good sign that this transposon promoter is also in innubila
- Looking closer at the BLAST hits, the largest one is is 78bp with a lot of miss-matches, ones that have less differences are more like ~59bp. The whole promoter is 280bp
- The longest “match” is 27,025,970 - 27,025,888 on the 2L chromosome. When looking at this region, there are no predicted genes around there. Without any other BLAST matches to anywhere near there, it’s really hard to guess if there is a promoter somewhere in there. For now I am going to give up on this promoter
- U6
- The top matches are for the X chromosome and the 3R chromosome. It’s on the 3R chromosome in dmel
- Again this is not good, there are only small matches. The longest match is about 56 bases but it’s not a very good one. The dmel promoter length is 421 bases. Again none of the separate matches are close to each other on the chromosomes
- I am not sure this is the right way to go about this. It might be helpful to find the U6 gene in innubila and select a similar region around it that the dmel promoter uses
- Ubiquitin/poly-ubiquitin
- This matches to 3L and 2L. Because it is on 3L in dmel, I’ll consider those matches
- The long match on the 3L is 142 bases long with a lot of missmatches, from 6336331 to 6336189
- The shorter 3L match is 38 bases long and from 6334434 to 6334395. This one says it’s near the feature “low quality protein ubiquitin c”
- These two regions are pretty close to each other so they might be close enough to be apart of the same promoter region
- The putative ubiquitin gene in innubila (LOC117788806) has good BLAST hits to dmel ubiquitin-63e and ubiquitin-5e
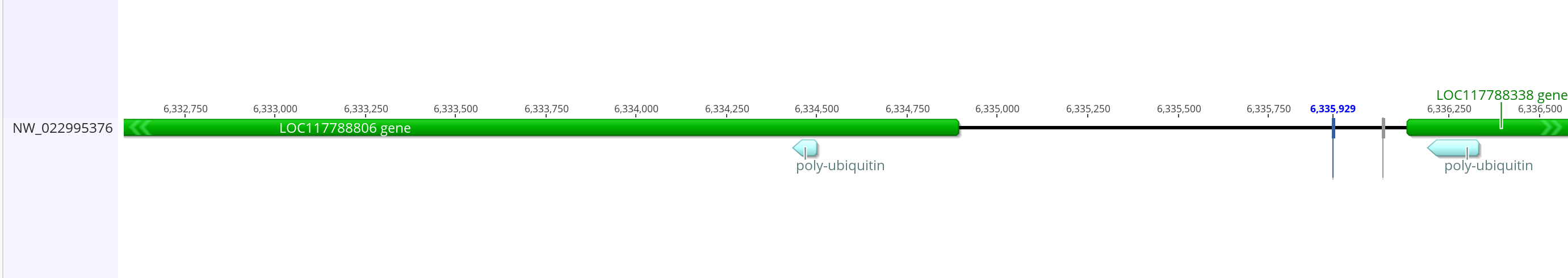
- The dmel promoter overlaps ~1,400 bases into the ubiquitin gene and ~150 bases in the gene next to it, and the whole promoter is 2,008 bases long. The two BLASTd regions are too far apart to match that exactly with innubila but I can do something similar
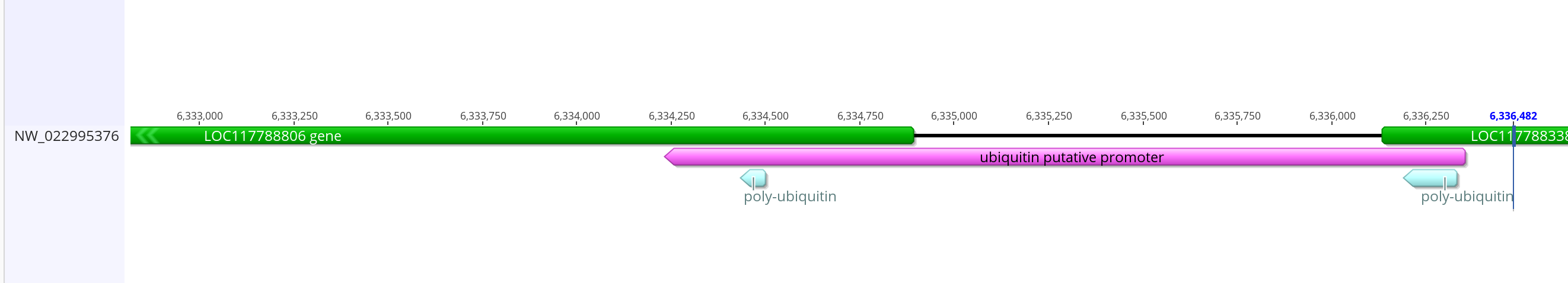
- This is how it compares with the mRNA from the gene
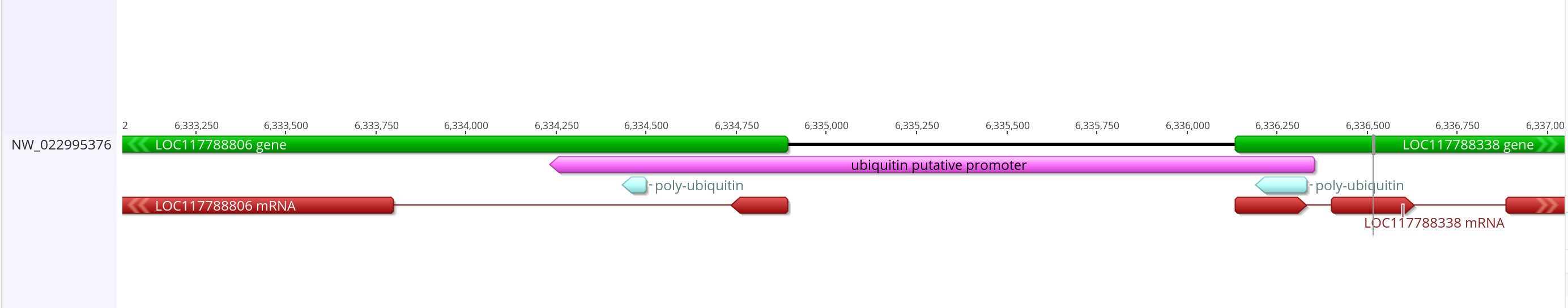
- Sequence link
- This putative promoter is 2,121 bases long
- Tubulin
- Matches to all chromosomes except for 4, but the best match is 3R which is where it is on dmel
- On 3R there are some long matches that look promising. There is a 360 base match from 24390062 to 24389703. Then there is a 109 base match from 24387521 to 24387408. There is a 139 base match from 24389494 to 24389355. And there is a 152 base match from 24389288 to 24389139. These are all in the same region of the chromosome, and some of them say they are near the tubulin alpha-1 chain feature
- In the innubila genome, these are upstream or overlapping with LOC117793066 which when BLASTd to dmel matches very well with alpha-tubulin 84b and 85e
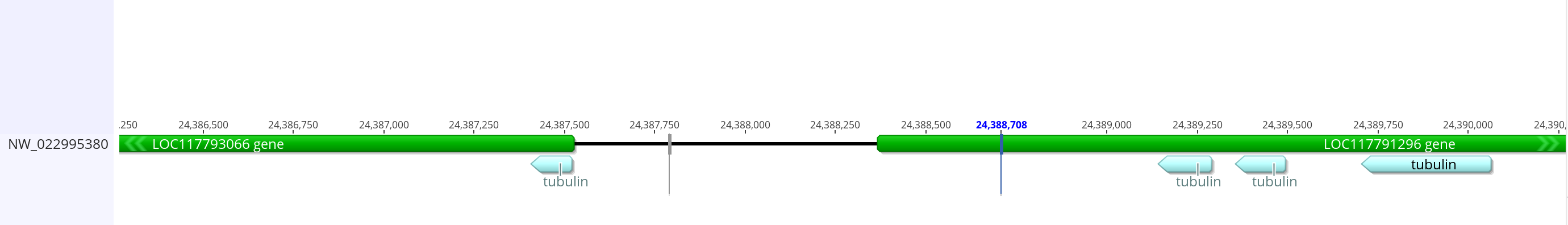
- The promoter from dmel is 2,399 bases long, and overlaps heavily with the gene before it and a small amount with the tubulin gene. I can do that with the innubila genome, but the promoter will be longer than the dmel one if I keep in all the BLAST matching sequences. I think it’s important to keep those sequences in the promoter because they seem to be conserved for some reason
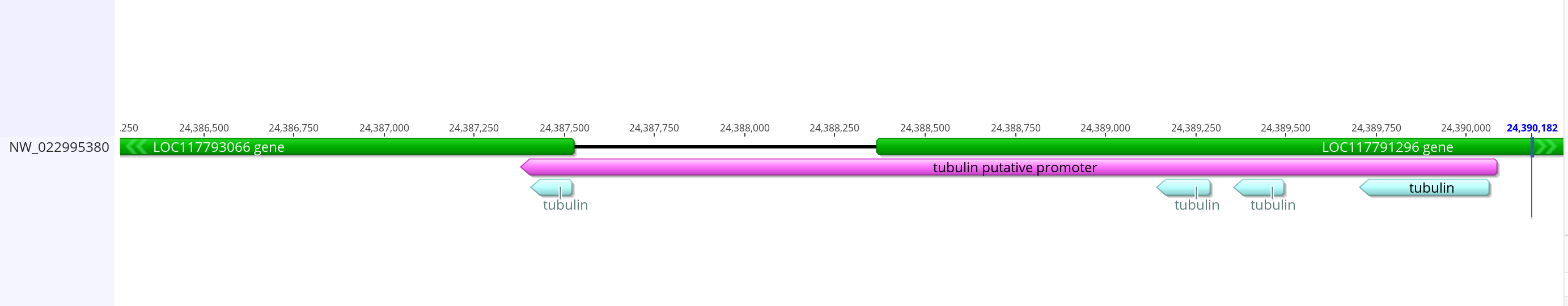
- This is how it compares with the mRNA from the gene
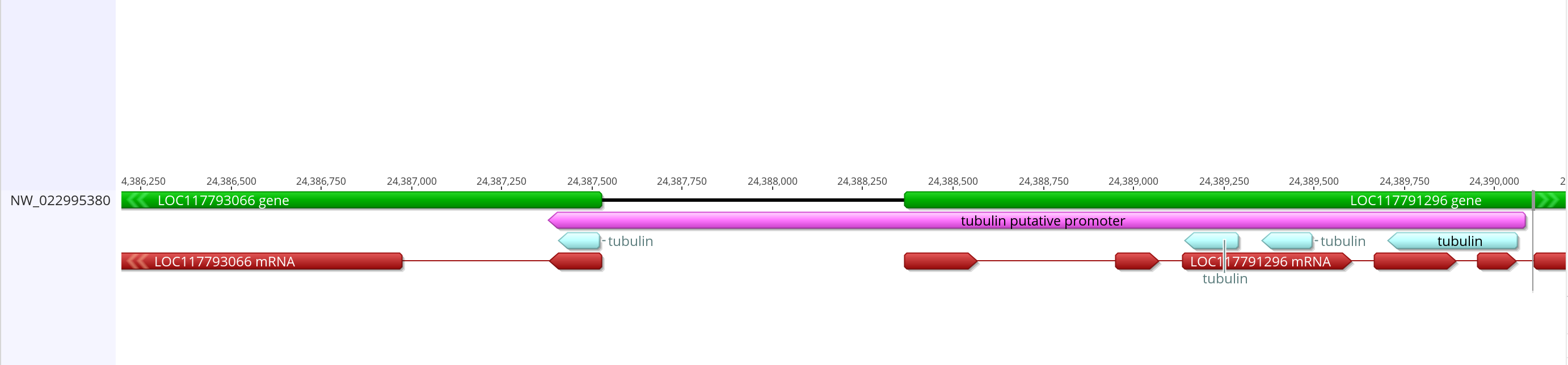
- Sequence link
- This putative promoter is 2,706 bases long
- Ef-1-alpha
- This had a couple of matches, but the strongest one is to 2R, which is where this gene is in dmel so I’m going to look here
- The longest match is ~90 bases from 3089592 to 3089504 on the 2R chromosome. There are a few other potentially closes matches on 2R as well. About 85 bases match from 3090707 to 3090617, and about 35 bases match from 3089730 to 3089697
- These also have the feature designation of elongation factor 1-alpha-1, so this seems to be in the right place. That feature is LOC117783331, which overlaps with all of the BLAST matching promoter sequences
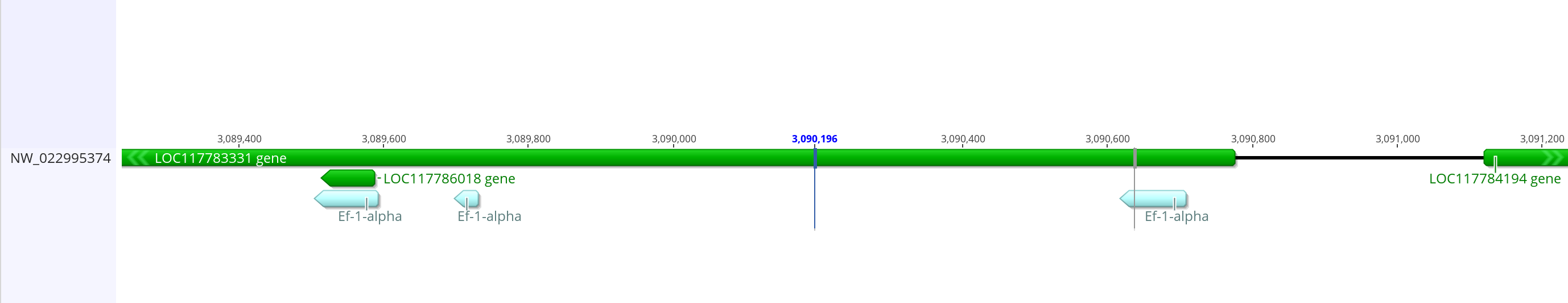
- The promoter from dmel is 2,315 bases long and completely overlaps with the Ef-1-alpha gene. The region including the BLAST matching sequences is only ~1,200 bases. If I actually go ~2,300 bases in the LOC117783331, it starts getting into the coding sequence/mRNA. I think that’s probably not needed so I’m going to make this putative promoter less than the dmel one
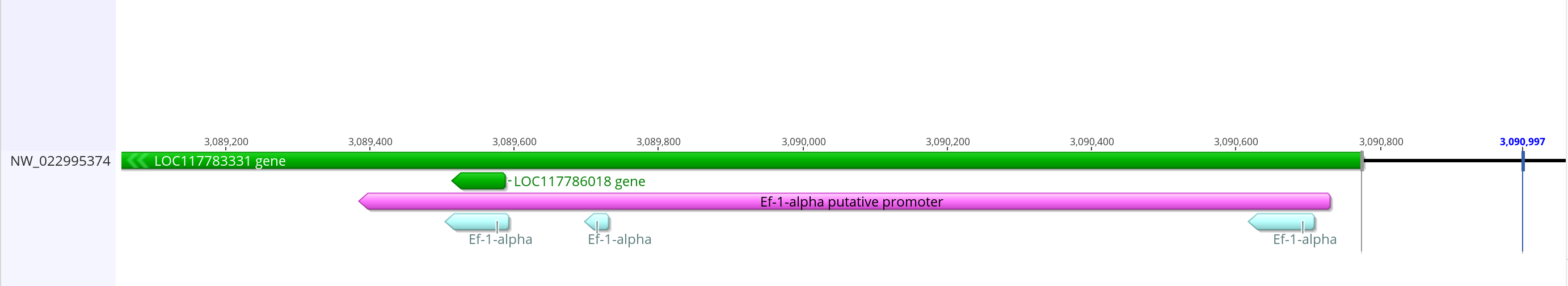
- Promoter sequence with mRNA prediction
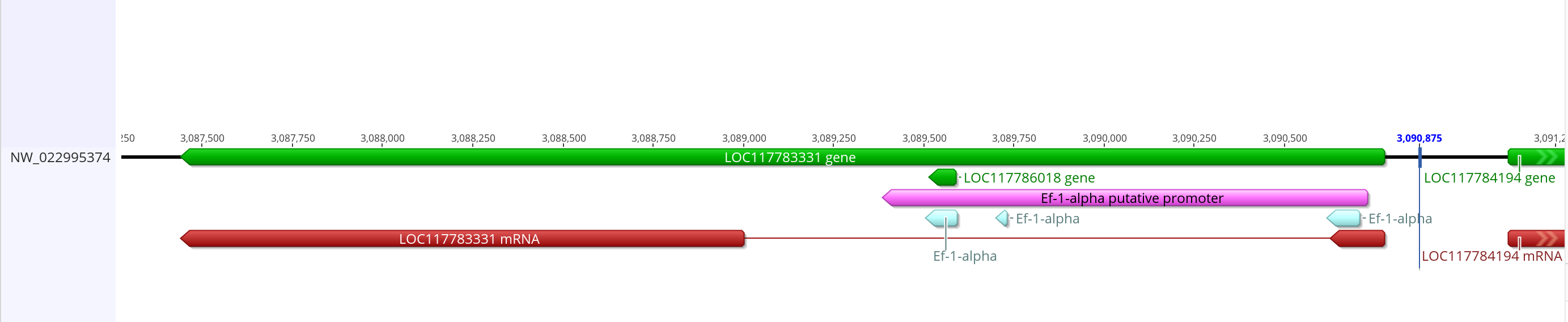
- Sequence link
- This putative promoter is 1,345 bases long