Using Qiagen Plasmid Midi Prep Kit to Extract pBeloBAC11 Using 150mL Overnight Culture
Notes
- Used 150mL of LB, used the same streaked plate as previous two attempts to pick a colony
- Separated out the volume into 3 50mL tubes through going through the genomic tip column, where the volumes were combined and ran through together
20220214 Making LB and Overnight Culture
- Prepared 250mL of LB
- 5g LB broth base in 250mL of distilled water, shaken until homogenous
- Aliquoted 150mL of LB into a 1 liter flask
- Covered the flask with foil
- Autoclaved in program 3
- Put flasks in the fridge after
- An hour later, placed the flask on the bench for a while to warm (around noon)
- Labeled the flask
- Started the bunsen burner
- Added 45ul of 50mg/mL of Chloramphenicol to the flask
- Picked 1 colony from the pBeloBAC11 streak plate with a p2 pipette tip, and dropped the tip into the flask
- Covered the flask with the same foil and placed in the 37 degree C shaking incubator overnight
20220215 MidiPrep Extraction
- Transferred volume from flask into 3 50mL conicals (about 45mL each)
- Took upstairs to the Egan Lab and centrifuged 6,000rcf, 4 degrees C, 20 minutes
- Got ice and put buffer P3 on ice to chill
- Prepared buffer P1
- Made an aliquot of 12mL of P1
- Added 12ul of 100mg/ul RNase A to the aliquot note, this was half the amount of RNase I was supposed to use, so there may be some RNA in the sample
- Kept the tube on ice
- Removed supernatant from the bacterial pellets after centrifugation
- Added 4mL of buffer P1 to each tube and vortexed until there was no longer a pellet or any clumps
- Added 4mL of buffer P2 to each tube
- Liquid became blue and the tubes were inverted to mix until all the liquid was blue and there were no longer any clumps - very viscous
- Tubes were incubated at room temp for 5 minutes
- Added 4mL of chilled buffer P3 to each tube
- Inverted to mix tubes until there was no longer any blue and any viscosity
- Lots of white precipitate showed up in the liquid
- Placed the tubes in the ice for 15 minutes
- Took the tubes upstairs to the Egan lab and centrifuged 40 min at 12,400rcf and 4 degrees C
- After: brought new 15mL tubes upstairs and a p1000 pipette and tips
- Beside the centrifuge, pipetted the supernatant off into new 15mL tubes for each tube (there was a huge white pellet)
- Centrifuged the supernatant tubes for 30 minutes at 12,400rcf and 4 degrees C
- While that was going, made a new conical of 100% isopropanol and 70% ethanol
- Set up one genomic tips over 50mL conical
- Added 4mL of buffer QBT to the tip and let it drip through to equilibrate
- Warmed the incubator to 65 degrees C and put buffer QF inside to warm
- Removed tubes from the centrifuge and transferred the supernatant to one new 50mL tube
- Transferred the supernatant liquid to the tip and let the liquid drip through
- This was done ~3 times and the waste conical was changed in between
- Transferred to a new 50mL conical for waste
- Added 12mL of buffer QC to the tip and let drip through
- Added another 12mL of buffer QC to the tip and let drip through
- Transferred the tip to new 15mL tube labeled as final tube
- Added 5mL of warmed buffer QF to the tip and let drip
- Added 3.92mL of 100% isopropanol to each tube and inverted to mix (tubes had ~5.6mL volume, 0.7 volumes is 3.92mL)
- Centrifgued tubes for 45 minutes at 12,400rcf at 4 degrees C. Made sure they were facing a certain way to look for the pellet
- Looked for pellets afterwards - pretty sure that these clear streaks are the pellet, they went away after hydration
- Decanted the supernatant next to the centrifuge in the Egan lab
- Added 2mL of 70% ethanol to each tube
- Placed tubes back in the centrifuge for 30 min, 12,400rcf at 4 degrees C, same tube orientation
- Poured off the ethanol and let the tubes air dry for ~10 minutes
- Added 200ul of Qiagen elution buffer (10mM Tris HCl pH 8.5) to where I thought the pellet might be, and tried to keep the liquid bubble on those streaks
- Placed the tubes on their sides in the 65 degree incubator for ~1hr
- Qubit: 48.1 ng/ul. That’s a total yield of ~9,620ng! I need 5ug for Genewiz so this should be enough
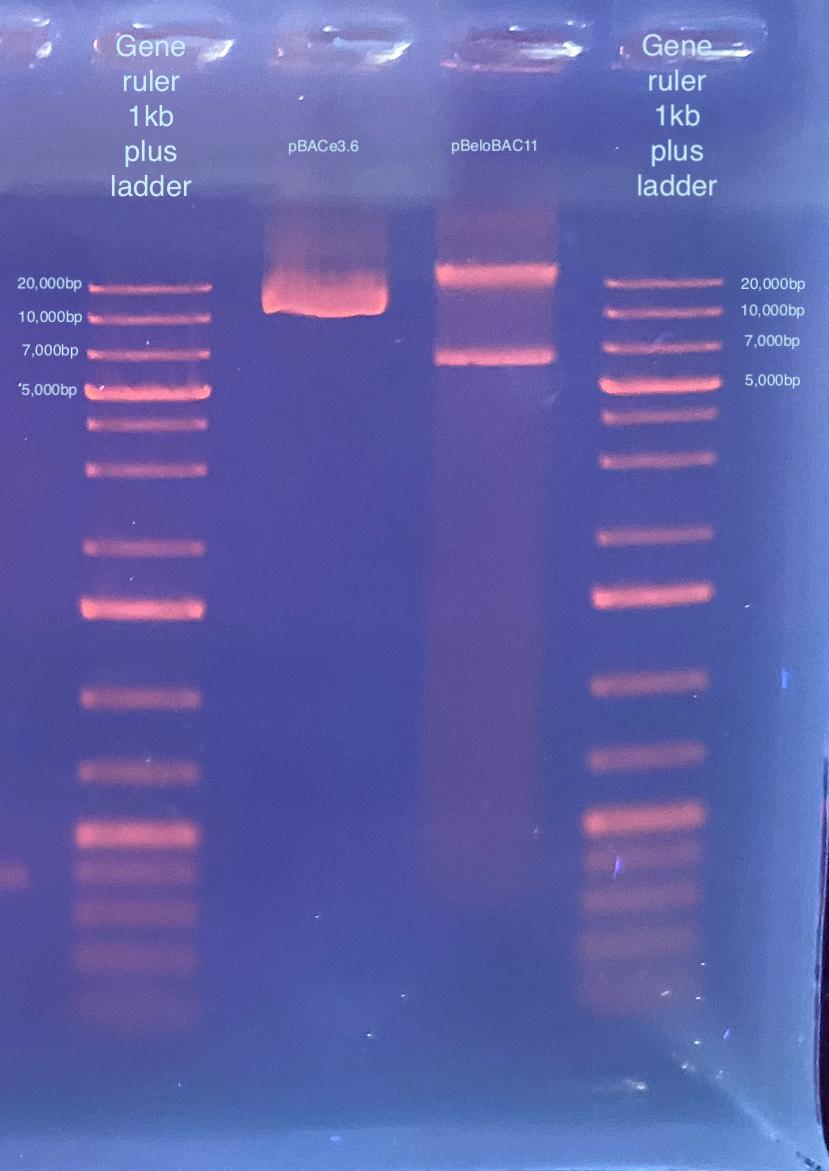
- Then I wanted to gel my two BAC extractions because they should have an expected size.
- pBACe3.6 should be 11.6kb in size
- pBeloBAC11 should be 7.5kb in size
- I took 3ul from each and ran on a 1% gel at 100V for 35 minutes:
I’m not entirely sure what to make of this gel, these are not exactly the right sizes. There also seems to be maybe some genomic DNA contamination. However these are circular so they may not run through the gel like a linear fragment might.
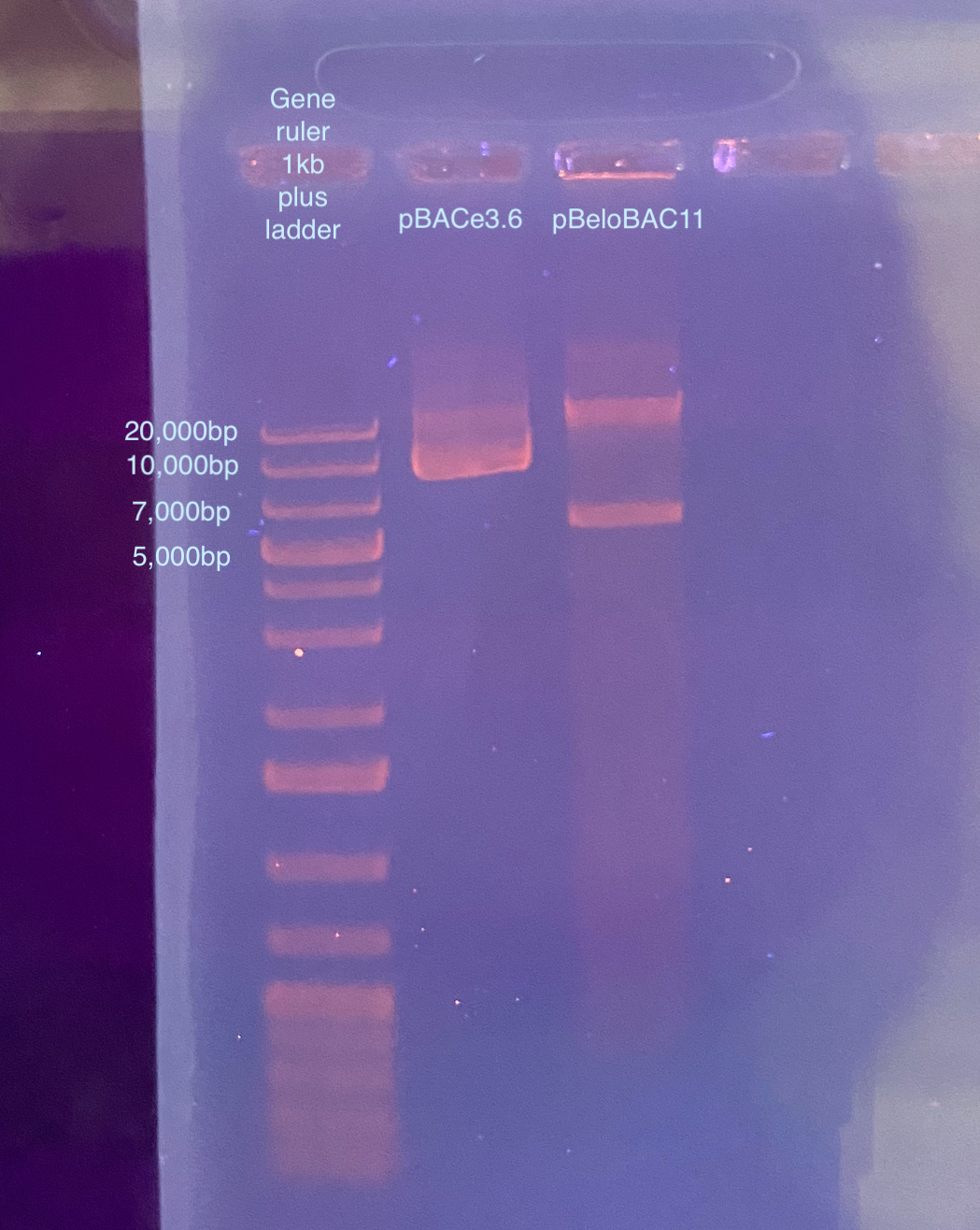
I ran another gel with two ladders and this shows more accurate sizes, however there still looks like there are 2 bands, particularly for pBeloBAC11.