Thoughts on Using 2 Cas9 Nickases on to Make the DiNV BAC
Background: Through reading of the Addgene CRISPR handbook, the IDT Nickase webpage, and the IDT Nickase Application Note
- There are two different mutations in the Cas9 protein, each produces an enzyme that only makes a 1 strand break
- D10A only cleaves the target strand
- H840A only cleaves the non-target strand
- Need two guide RNAs, one for each strand/each Cas9. You need two Cas9s to make two cuts on the DNA to effectively create a DSB
- The D10A Cas9 has a higher editing efficiency and works better to generate homology directed repair, so we should use that one
- The orientation of sgRNAs that works best is “PAM-out”, where the PAM sites are on the outside of the two sgRNAs (see image)
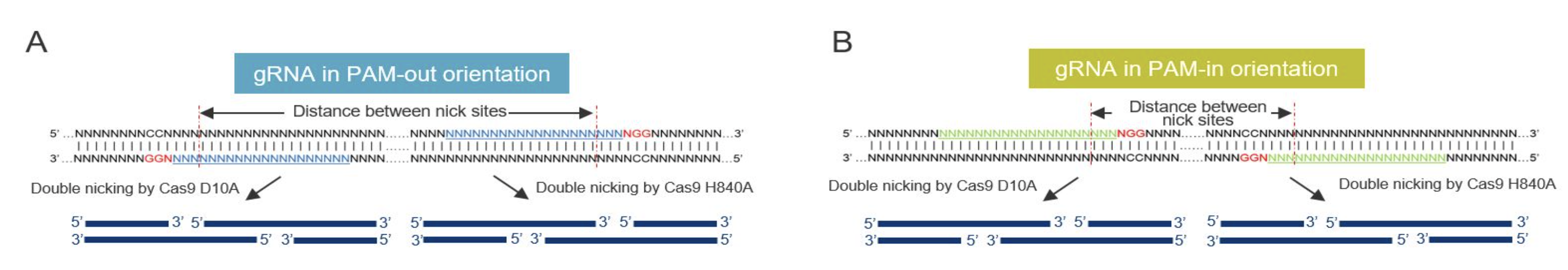
- The ideal number of bases between nick sites (so this is not between sgRNAs, but their specific nick sites) is 37-68bp
First I wanted to see if it was possible with our 4 regions of the DiNV genome to get 2 sgRNAs that have opposite orientation within these distances
77kb Region
- 85 and 31
- 85 cuts at 77,497
- 31 cuts at 77,540
- Nick distance is 43 bp
- 85 activity score: 0.504
- 31 activity score: 0.545
- 0 off-tartget in CDS for either
- 89 and 27
- 89 cuts at 77,350
- 27 cuts at 77,400
- Nick distance is 50bp
- 89 activity score: 0.351
- 27 activity score: 0.716
- 0 off-tartget in CDS for either
44kb Region
- 58 and 1
- 58 cuts at 44,346
- 1 cuts at 44,395
- Nick distance is 49bp
- 58 activity score: 0.545
- 1 activity score: 0.353
- 7.31% off-target score for guide 1
- 54 and 3
- 54 cuts at 44,450
- 3 cuts at 44,512
- Nick distance is 62bp
- 54 activity score: 0.662
- 3 activity score: 0.516
- 0 off-target in CDS for either
- 44 and 11
- 44 cuts at 44,882
- 11 cuts at 44,948
- Nick distance is 66bp
- 44 activity score: 0.667
- 11 activity score: 0.332
- 0 off-target in CDS for either
- 44 and 10
- 44 cuts at 44,882
- 10 cuts at 44,926
- Nick distance is 44bp
- 44 activity score: 0.667
- 10 activity score: 0.340
- 0 off-target in CDS for either
- 32 and 14
- 32 cuts at 45,061
- 14 cuts at 45,119
- Nick distance is 59bp
- 32 activity score: 0.559
- 14 activity score: 0.370
- 0 off-target in CDS for either
- 30 and 15
- 30 cuts at 45,090
- 15 cuts at 45,150
- Nick distance is 60bp
- 30 activity score: 0.589
- 15 activity score: 0.509
- 0 off-target in CDS for either
- 26 and 18
- 26 cuts at 45,267
- 18 cuts at 45,332
- Nick distance is 65bp
- 26 activity score: 0.533
- 18 activity score: 0.338
- 0 off-target in CDS for either
75kb Region
- 81 and 2
- 81 cuts at 74,550
- 2 cuts at 74,604
- Nick distance is 54bp
- 81 activity score: 0.586
- 2 activity score: 0.555
- 0 off-target in CDS for either
- 81 and 1
- 81 cuts at 74,550
- 1 cuts at 74,603
- Nick distance is 53bp
- 81 activity score: 0.586
- 1 activity score: 0.583
- 0 off-target in CDS for either
- 71 and 37
- 71 cuts at 75,338
- 37 cuts at 75,278
- Nick distance is 55bp
- 71 activity score: 0.572
- 37 activity score: 0.578
- 0 off-target in CDS for either
- 68 and 42
- 68 cuts at 75,338
- 42 cuts at 75,393
- Nick distance is 55bp
- 68 activity score: 0.504
- 42 activity score: 0.718
- 0 off-target in CDS for either
- 64 and 43
- 64 cuts at 75,427
- 43 cuts at 75,479
- Nick distance: 52bp
- 64 activity score: 0.662
- 43 activity score: 0.509
- 0 off-target in CDS for either
- 57 and 51
- 57 cuts at 75,540
- 51 cuts at 75,593
- Nick distance is 53bp
- 57 activity score: 0.316
- 51 activity score: 0.675
- 0 off-target in CDS for either
130kb Region
- 64 and 30
- 64 cuts at 129,798
- 30 cuts at 129,850
- Nick distance is 52bp
- 64 activity score: 0.552
- 30 activity score: 0.589
- 0 off-target in CDS for either
- 9 and 28
- 9 cuts at 130,765
- 28 cuts at 130,835
- Nick distance is 70bp
- 9 activity score: 0.585
- 28 activity score: 0.624
- Both have off-target hits in the DiNV genome
I think it’s best not to pick any that have off-target hits. Or probably one of the guides with an activity score below 0.5. This leaves about 10 pairs of sgRNAs left.