Using Qiagen Plasmid Midi Prep Kit to Extract pBeloBAC11 and pBACe3.6
Notes
- Goal is to extract the BACs from their E. coli hosts and use them to send to Genewiz to insert our homology arms into
- I used only 4mL of culture for the extraction, I think I should have used way more. Looking at the protocol it says the max volume is 25mL for a high copy plasmid, and in Mason’s notebook he said he used 50mL. I will probably want to try again doing maybe 20mL
- I used the Egan lab centrifuge on the 8th floor. It’s maximum speed is 15,000rcf, but the protocol says to use at least 20,000rcf for many spins. Unfortunately the tubes we have are only rated to 10,400rcf! So that’s the fastest I went. I got some new tubes at the Biostore that say they can go 12,500rcf so I will use those next time
- The Egan lab centrifuge is a fixed angle rotor, so I’m not sure exactly where the pellet of DNA should have been
- Every centrifuge I added on at least 10 more minutes to the spin because of the slower speeds and that is what Mason said in his notebook that he did
- Mason also resuspended his DNA for 1 hour at 65 degrees so I did that too
- I also left the DNA out on the bench overnight to potentially resuspend more
20220207 Plating BACs on LB
- Warmed two LB plates in the 37 degree incubator
- Took both BAC glycerol stocks from the -80 and kept on ice for use (pBACe3.6 and pBeloBAC11)
- Made a diluted stock of chloramphenicol to add 20ug/mL to each plate:
- 15ul of 50mg/mL of stock
- 15ul of 100% ethanol
- Turned on the bunsen burner
- Make a sterile spreader with a Pasteur pipette
- Added 15ul of diluted chloramphenicol to each plate and quickly spread the antibiotic with the glass spreader
- Used a plastic loop for each BAC, and took a small amount of glycerol stock and spread on the LB plates
- Labeled plates and placed upside down in the 37 degree incubator overnight
20220208 Making Overnight Cultures
- Make 4 test tubes, each with 4mL of LB (combined 2 of the already made 2mL ones)
- Labeled 2 for each BAC
- Turned on bunsen burner
- Added 1.2ul of 50mg/ul of chloramphenicol to each test tube (to 20ug/mL conc)
- Took a p2 pipette tip and used it to pick 1 colony and added it to the appropriate test tube. Repeated for other test tubes (each BAC gets 2 tubes)
- Placed tubes in the 37 degree C incubator shaking
- Parafilmed up the plates and put in the fridge in 4012
20220209 Extraction
- Texted member of the Egan lab to say I would be up in ~10 minutes so they could cool down the centrifuge
- Transferred the liquid in 2 of the test tubes of the overnight cultures (the other two were backups) into respectively labeled 15mL tubes
- Centrifuged the 15mL tubes at 6000rcf for 25 minutes at 4 degrees C in the Egan lab
- Got ice and put buffer P3 on ice to chill
- Prepared buffer P1
- Added the entire volume of Lyse Blue reagent to the entire P1 bottle (kit had not been opened) and vigorously shook the bottle
- Made an aliquot of 8mL of P1
- Added 16ul of 100mg/ul RNase A to the aliquot (for a final concentration of 100ug/mL)
- Kept the tube on ice
- Removed supernatant from the bacterial pellets after centrifugation
- Added 4mL of buffer P1 to each tube and vortexed until there was no longer a pellet or any clumps
- Added 4mL of buffer P2 to each tube
- Liquid became blue and the tubes were inverted to mix until all the liquid was blue
- Tubes were incubated at room temp for 5 minutes
- Added 4mL of chilled buffer P3 to each tube
- Inverted to mix tubes until there was no longer any blue
- Lots of white precipitate showed up in the liquid
- Placed the tubes in the ice for 15 minutes
- Took the tubes upstairs to the Egan lab and centrifuged 40 min at 10,400rcf and 4 degrees C
- After: brought new 15mL tubes upstairs and a p1000 pipette and tips
- Beside the centrifuge, pipetted the supernatant off into new 15mL tubes for each tube (there was a huge white pellet that was so much it went all the way up the side of the tube)
- Centrifuged the supernatant tubes for 30 minutes at 10,400rcf and 4 degrees C
- While that was going, made a new conical of 100% isopropanol and 70% ethanol
- Set up two genomic tips over 50mL conicals and labeled with tape
- Added 4mL of buffer QBT to each tip and let it drip through to equilibrate
- Warmed the incubator to 65 degrees C and put buffer QF inside to warm
- Removed tubes from the centrifuge and transferred the supernatant to new 15mL tubes (there was hardly any pellet this time)
- Transferred the supernatant liquid to their respective tips and let the liquid drip through
- Transferred to a new 50mL conical for waste
- Added 10mL of buffer QC to each tip and let drip through
- Added another 10mL of buffer QC to each tip and let drip through
- Transferred the tips to new 15mL tubes labeled as final tubes
- Added 5mL of warmed buffer QF to each tip and let drip
- Added 3.85mL of 100% isopropanol to each tube and inverted to mix (tubes had ~5.5mL volume, 0.7 volumes is 3.85mL)
- Centrifgued tubes for 45 minutes at 10,400rcf at 4 degrees C. Made sure they were facing a certain way to look for the pellet
- Looked for pellets afterwards - not really sure what to look for. The protocol says the pellet should be “glassy.” Additionally because the centrifuge is fixed angle, it’s not gong to be at the base of the tube. I saw these streaks of clear something on each of the tubes. But the more I look at them the more I think they are stress cracks in the tube bc it was at its maximum speed
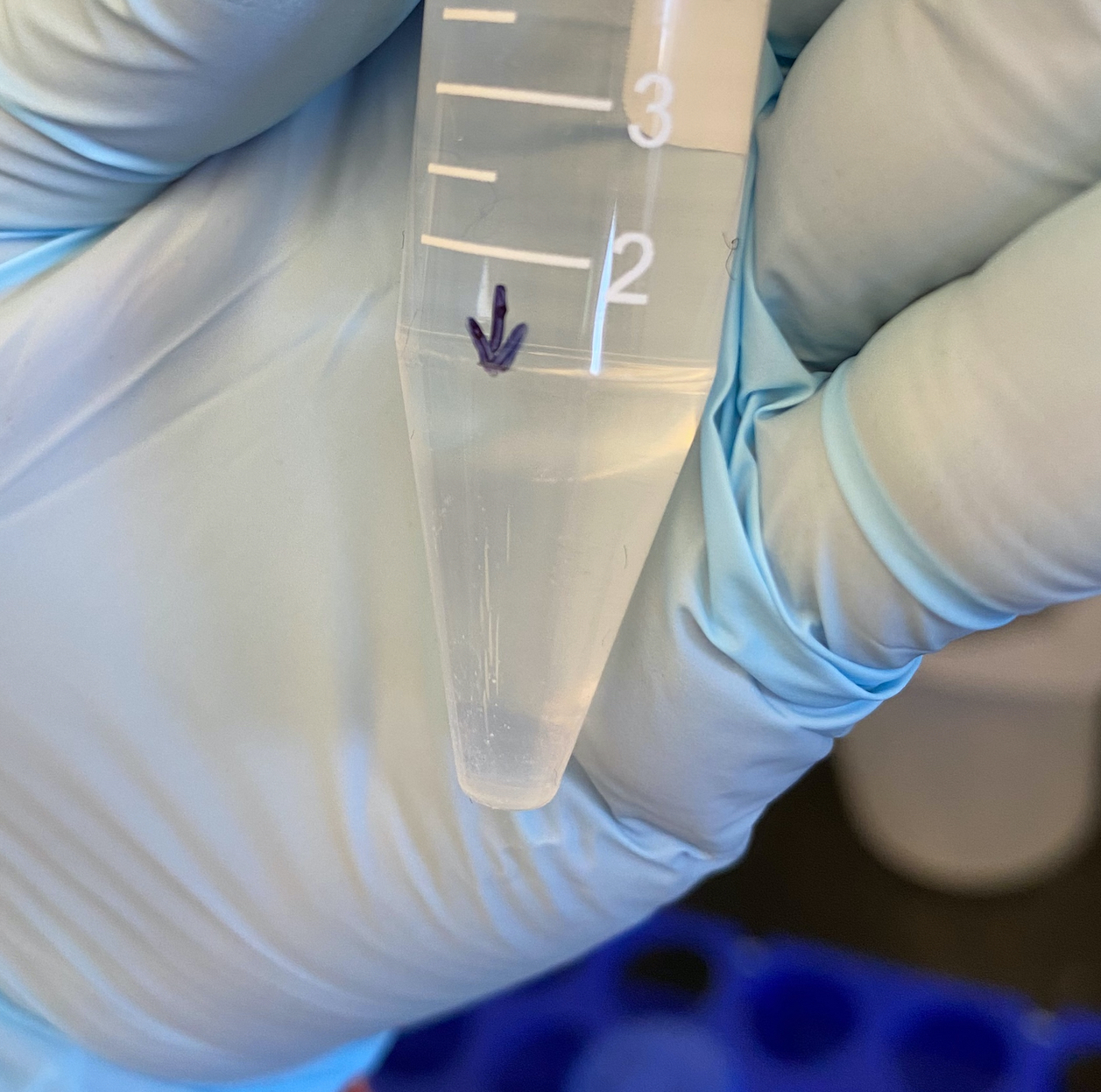
- Decanted the supernatant next to the centrifuge in the Egan lab
- Added 2mL of 70% ethanol to each tube
- Placed tubes back in the centrifuge for 30 min, 10,400rcf at 4 degrees C, same tube orientation
- Poured off the ethanol and let the tubes air dry for ~10 minutes
- Added 100ul of Qiagen elution buffer (10mM Tris HCl pH 8.5) to where I thought the pellet might be, somewhat on the side of the conical part and tapped the liquid bubble around that area for a few minutes
- Placed the tubes on their sides in the 65 degree incubator for ~1hr
- Qubit after 1hr:
- pBeloBAC11: 0.918ng/ul
- pBACe3.6: 22.3ng/ul
- I wasn’t sure if they had completely resuspended or not so I left them on the bench until the next morning where I qubited them again
- Thursday AM Qubit:
- pBeloBAC11: 1.11ng/ul
- pBACe3.6: 23.4ng/ul