20211213 Re-PCR RPL11 and p47 Testing Diluting DNA 1:10 and Doubling DNA Input
| Sample |
DNA |
Molcular grade water |
| 3MLB-150 |
1 |
9 |
| 3mLB-UP |
1 |
9 |
| 3mLB-LOW |
1 |
9 |
- Make RPL11 master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM vir_RPL11_F primer * 8.8 = 2.2ul
- 0.25ul 10uM vir_RPL11_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 0.4ul molecular grade water * 8.8 = 3.52ul
- Added master mix, DNA, and water to strip tubes. Samples with “2” next to them had twice the amount of DNA added to them (~16-18ng total), and samples with the “D” were the 1:10 diluted samples
| tube # |
sample |
ul RPL11 MM |
ul DNA |
ul water |
| 1 |
3mLB-150-2 |
4 |
2 |
4 |
| 2 |
3mLB-UP-2 |
4 |
6 |
0 |
| 3 |
2mLB-LOW-2 |
4 |
4 |
2 |
| 4 |
3mLB-150-D |
4 |
1 |
5 |
| 5 |
3mLB-UP-D |
4 |
3 |
3 |
| 6 |
3mLB-LOW-D |
4 |
2 |
4 |
| 7 |
neg control |
4 |
0 |
6 |
| 8 |
pos control |
4 |
1 |
5 |
- Put in PCR program and geled the next day
- Made p47 master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM p47_F primer * 8.8 = 2.2ul
- 0.25ul 10uM p47_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 0.4ul molecular grade water * 8.8 = 3.52ul
- Added master mix, DNA, and water to strip tubes. Samples with “2” next to them had twice the amount of DNA added to them (~16-18ng total), and samples with the “D” were the 1:10 diluted samples (0.8-0.9ng)
| tube # |
sample |
ul p47 MM |
ul DNA |
ul water |
| 1 |
3mLB-150-2 |
4 |
2 |
4 |
| 2 |
3mLB-UP-2 |
4 |
6 |
0 |
| 3 |
2mLB-LOW-2 |
4 |
4 |
2 |
| 4 |
3mLB-150-D |
4 |
1 |
5 |
| 5 |
3mLB-UP-D |
4 |
3 |
3 |
| 6 |
3mLB-LOW-D |
4 |
2 |
4 |
| 7 |
neg control |
4 |
0 |
6 |
| 8 |
pos control |
4 |
1 |
5 |
- Put in PCR program and geled the next day
- Next day a 1% gel was run for ~30 minutes at 90V
- This gel came out really fuzzy and hard to read, it is not clear if there is primer dimer for RPL11 or actual amplification. I decided after this to go forward only troubleshooting the RPL11 amplification and also to run all my gels on 2% agarose to actually separate out something so small like primer dimer and a 137bp product

20211214 RPL11 Diluted PCR with 2% Gel
- Tried doing the diluted DNA and then also doubling the amount of the diluted DNA
- Made master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM vir_RPL11_F primer * 8.8 = 2.2ul
- 0.25ul 10uM vir_RPL11_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 0.4ul molecular grade water * 8.8 = 3.52ul
- Added master mix, DNA, and water to strip tubes. Samples with “D-2” next to them had twice the amount of diluted DNA added to them (~1.6-1.8ng total), and samples with the “D” were the 1:10 diluted samples (0.8-0.9ng)
| tube # |
sample |
ul RPL11 MM |
ul DNA |
ul water |
| 1 |
3mLB-150-D |
4 |
2 |
4 |
| 2 |
3mLB-UP-D |
4 |
6 |
0 |
| 3 |
2mLB-LOW-D |
4 |
4 |
2 |
| 4 |
3mLB-150-2 |
4 |
1 |
5 |
| 5 |
3mLB-UP-2 |
4 |
3 |
3 |
| 6 |
3mLB-LOW-2 |
4 |
2 |
4 |
| 7 |
neg control |
4 |
0 |
6 |
| 8 |
pos control |
4 |
1 |
5 |
- Put in PCR program and geled the next day
- Next day a 2% gel was run for ~30 minutes at 90V
- Gel came out bad, could see the positive control but nothing else. I also poured the gel way too thick
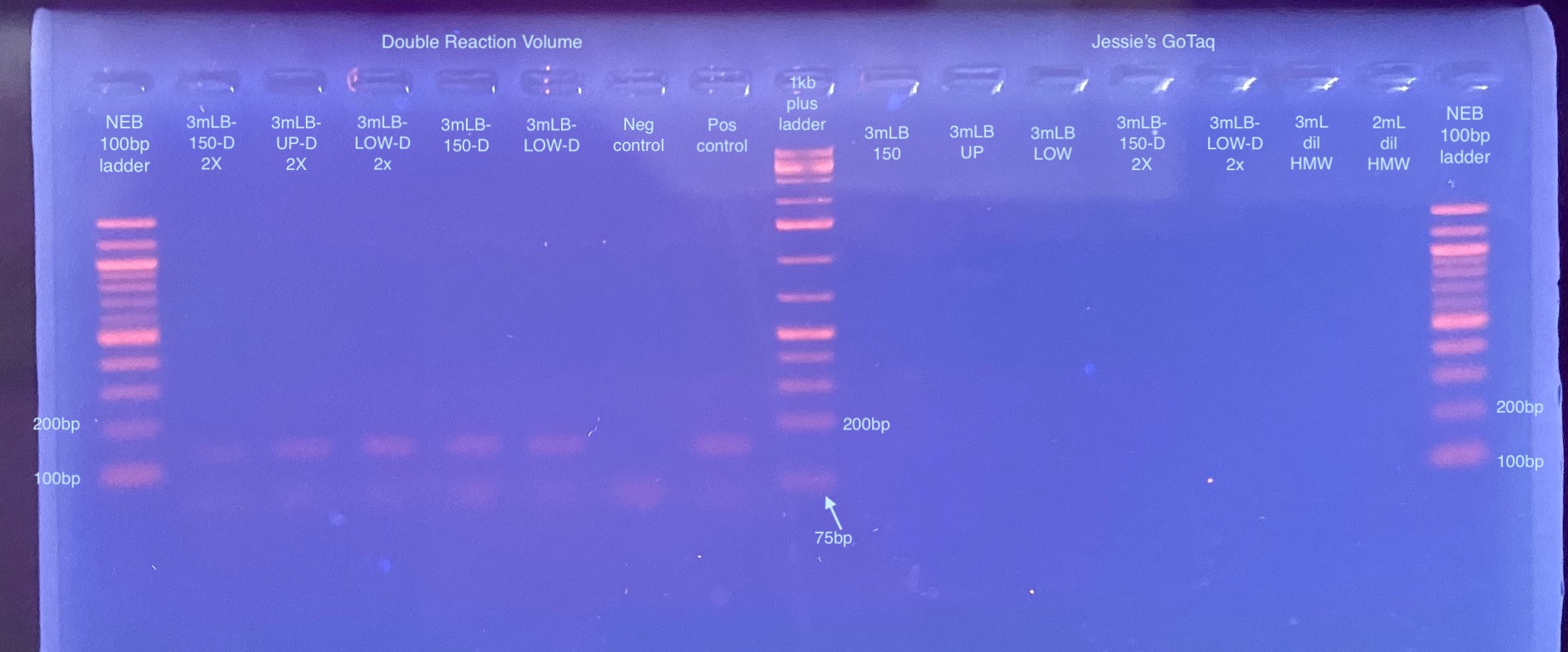
20211215 Testing RPL11 PCR With Double Reaction Volume, GoTaq, and With Reduced PCR time
- Wanted to try testing a 20ul PCR volume, GoTaq that Jessie said worked for her, and I wanted to modify the PCR times (reduce annealing and extension times) to see if they would work because the PCR program right now is very long
- Because the diluted gel extracted DNA is the only thing that maybe worked, that’s what I used for the doubled reaction volume. I did a mix of sample types for the GoTaq, and samples that I knew worked for testing the PCR conditions
- First I made the 2X reaction volume master mix:
- 2ul NEB buffer * 7.7 = 15.4ul
- 2ul dNTPs * 7.7 = 15.4ul
- 2ul MgCl2 * 7.7 = 15.4
- 0.5ul vir_RPL11_F primer * 7.7 = 3.85ul
- 0.5ul vir_RPL11_R primer * 7.7 = 3.85ul
- 0.2ul NEB Taq * 7.7 = 1.54ul
- 0.8ul molec grade water * 7.7 = 6.16ul
- Added master mix, DNA, and water to strip tubes. Samples with “D-2” next to them had twice the amount of diluted DNA added to them (~1.6-1.8ng total), and samples with the “D” were the 1:10 diluted samples (0.8-0.9ng)
- I’m not sure where my calculations got a little off, I had to add a lot of water in per sample to make the reaction volume 20ul per sample
| tube # |
sample |
ul RPL11 MM |
ul DNA |
ul water |
| 1 |
3mLB-150-D-2 |
8 |
2 |
10 |
| 2 |
3mLB-UP-D-2 |
8 |
6 |
6 |
| 3 |
2mLB-LOW-D-2 |
8 |
4 |
8 |
| 4 |
3mLB-150-D |
8 |
1 |
11 |
| 5 |
3mLB-LOW-D |
8 |
2 |
12 |
| 6 |
neg control |
8 |
0 |
12 |
| 7 |
pos control |
8 |
1 |
11 |
- Made master mix for GoTaq, 10ul reaction volume
- 5ul GoTaq (2X) * 10 = 50ul
- 4ul molecular grade water * 10 = 40ul
- Added master mix, DNA, and water to strip tubes
| tube # |
sample |
ul GoTaq MM |
ul DNA |
ul water |
| 8 |
3mLB-150 |
9 |
1 |
0 |
| 9 |
3mLB-UP |
9 |
1 |
0 |
| 10 |
3mLB-LOW |
9 |
1 |
0 |
| 11 |
3mLB-150-D |
9 |
1 |
0 |
| 12 |
3mLB-LOW-D |
9 |
1 |
0 |
| 13 |
3mL HMW |
9 |
1 |
0 |
| 14 |
2mL HMW |
9 |
1 |
0 |
| 15 |
neg control |
9 |
0 |
1 |
| 16 |
pos control |
9 |
1 |
0 |
- These were placed in the same PCR thermocycler with the regular PCR program
- 95 degrees C 3 minutes
- 95 degrees C 30 seconds
- 57 degrees C 1 minute
- 68 degrees C 1 minute 30 seconds
- 68 degrees C 5 minutes
- 12 degree C hold
- bold sections are cycled 34 times
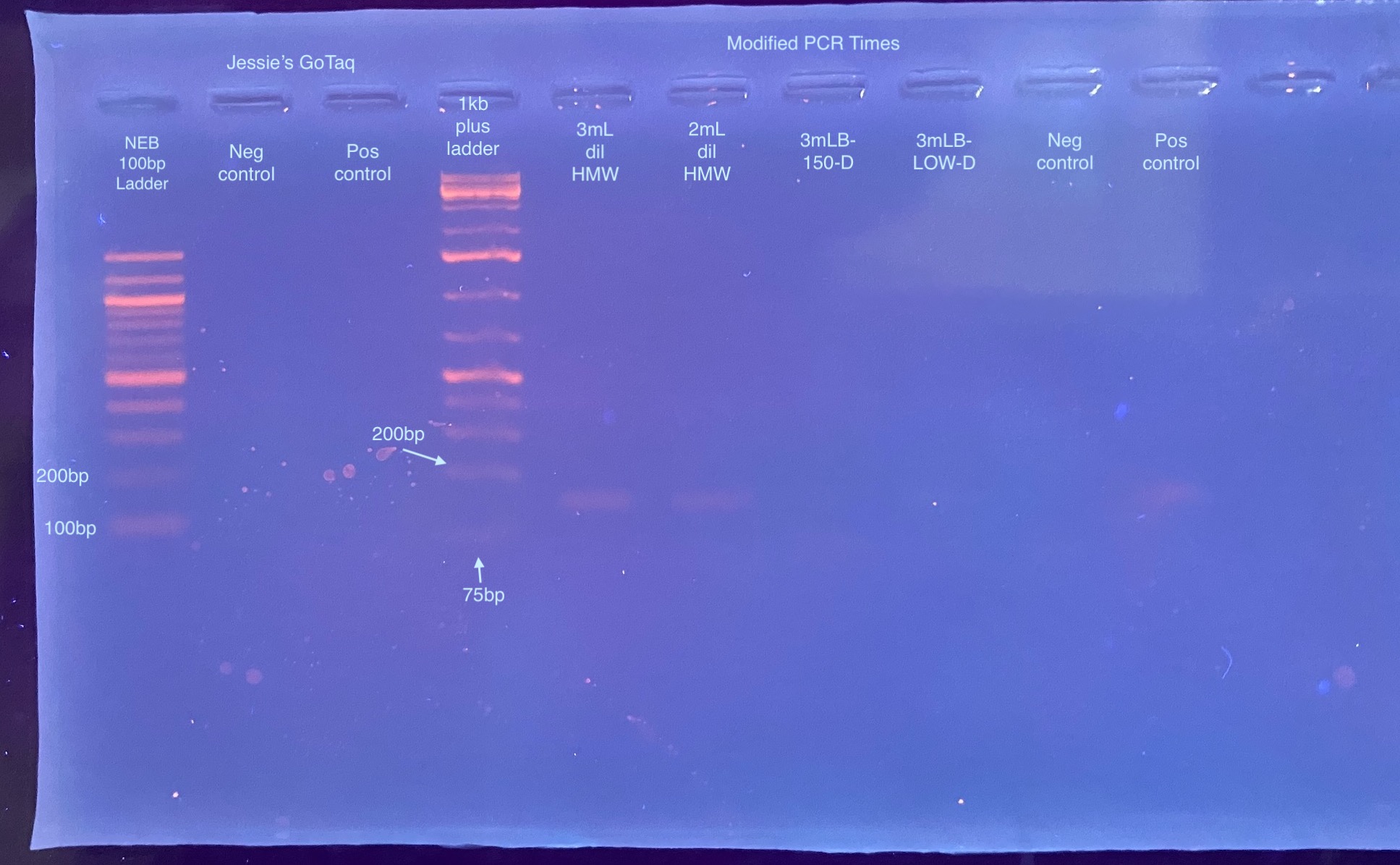
- Made a separate PCR reaction set up for testing different PCR program times
- Made master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM vir_RPL11_F primer * 8.8 = 2.2ul
- 0.25ul 10uM vir_RPL11_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 0.4ul molecular grade water * 8.8 = 3.52ul
- Added master mix, DNA, and water to strip tubes
| tube # |
sample |
ul RPL11 MM |
ul DNA |
ul water |
|
| 1 |
3mL |
8 |
1 |
1 |
|
| 2 |
2mL |
8 |
1 |
1 |
|
| 3 |
3mLB-150-D |
|
8 |
1 |
1 |
| 4 |
3mLB-LOW-D |
8 |
2 |
0 |
|
| 6 |
neg control |
8 |
0 |
2 |
|
| 7 |
pos control |
8 |
1 |
1 |
|
- Placed these in a slightly modified PCR:
- 95 degrees C 3 minutes
- 95 degrees C 30 seconds
- 57 degrees C 30 seconds
- 72 degrees C 1 minute
- 72 degrees C 5 minutes
- 12 degree C hold
- bold sections are cycled 34 times
- All of these were geled the next day on a 2% gel, poured low, and run for 30 minutes at 90V