Attempting Again the Genomic Tip Blood & Cell Culture DNA Midi Kit HMW Extraction on One Cell Control and One DiNV Positive Cell Culture Samples with a Faster Centrifuge
Notes
- Samples:
- Pos-2: DiNV GS 20181123 DV-1-1 day 10 lysate
- CC-2: DiNV GS 20181123 DV-1-CC day 10
- Using 600ul input this time to try to increase yield of DNA
- Going to let the pellet dry for longer (30 min if possible) after the ethanol wash because in the regular extraction protocol they let their pellets dry out for a lot longer. Apparently ethanol can inhibit DNA from going into solution
- Going to leave the samples resuspending for an extra day before trying the QC
- Trying a new centrifuge this time for the precipitation that goes to 10,000g. However it is a fixed angle rotor, so the pellet location may be up the side of the tube
- Used different 15mL tubes this time: thermofisher scientific 339651. These are rated to go 10,400g. I decided to use the centrifuge at 9,500g to be safe
20211101
Sample Washing
- Placed 1X PBS and molec grade water on ice
- Kept samples on ice
- Set centrifuge on the 5th floor to go to 4 degrees
- Inverted sample tubes before pipetting to mix up the cells: didn’t see any cell clumps here, but the liquid looked a little cloudy
- Clipped the ends of p1000 pipette tips for each sample
- Transferred 600ul of each sample into their own 15mL tube
- Added 9mL cold PBS to each tube
- Centrifuged tubes at 1,500rcf for 10 minutes at 4 degrees C
- Removed supernatant from each tube without disturbing the pellet
- There was a visible small pellet in each tube
- Added 10mL cold PBS to each tube
- Centrifuged tubes at 1,500rcf for 10 minutes at 4 degrees C
- Removed supernatant
- Added 2mL cold PBS to each tube
- Added 2mL cold buffer C1 to each tube
- Added 6mL cold molecular grade water to each tube
- Vortexed to get the pellet to resuspend
- Let tubes sit on ice for 10 minutes
- Centrifuged 1,300rcf for 15 minutes at 4 degrees C
- Removed supernatant without disturbing the pellet
- There is a small pellet in each sample
- Added 1mL cold buffer C1 to each tube
- Added 3mL cold molecular grade water to each tube
- Vortexed breifly
- Centrifuged 1,300rcf for 15 minutes at 4 degrees C
- Removed supernatant
Incubation
- Added 5mL buffer G2 to each tube
- Added 95mL proteinase K to each tube
- Vortexed tubes briefly
- I did see the pellet come up again and hopefully get fully mixed
- Placed tubes in the incubator at 50 degrees C for ~60 minutes
Genomic Tip Extraction
- Set up two tips over 50mL conicals
- Added 4mL buffer GBT to each tip and let drip
- Drip times were pretty consistent between the tips, the POS-2 sample tip was a little slower, but not by much. Max drip time was ~10 min which is great
- Vortexed sample tubes briefly
- Cut p1000 tips for each samples
- Added total volume (~5mL) of each sample to their respective tips and let drip
- Placed buffer QF in the incubator to warm to 50 degrees C
- Added 7.5mL of buffer QC to each tip
- Transferred tips to a new 15mL waste conical
- Added 7.5mL of buffer QC to each tip
- Transferred tips to new 15mL conicals labeled for final tubes
- Added 5mL of warmed buffer QF to each tip and let drip
Precipitation
- Added 3.8mL of 100% isopropanol to each elutent tube and inverted multiple times to mix
- I noticed here that the volumes in the elutent tubes were closer to 6mL than 5. The isopropanol volume is supposed to be 0.7 volumes of the elutent volume. The volumes were slightly different between the two samples and there wasn’t a way to measure without pipetting. So I guessed 3.8mL would be closer to being correct
- Centrifuged tubes in the Slusky Lab at 4 degrees C 9,500rcf for 30 min
- This was in a fixed angle rotor with a 23 degree angle, ideally we want a swing bucket but this may be the best we have access to. Rotor may not have been at 4 degrees when it started, but the centrifuge was at 4 degrees
- Afterwards it was very hard to see a pellet
- With the fixed angle rotor, the pellet is going to be on the side of the tube. I had situated each tube in the rotor so that the side with the white patch was facing the outside of the rotor so I would at least have an idea where the pellet should be. After the first spin I thought I saw a pellet very faintly on the side of the tube (see circle in image), however the tubes had condensation and that might have cause the smudge
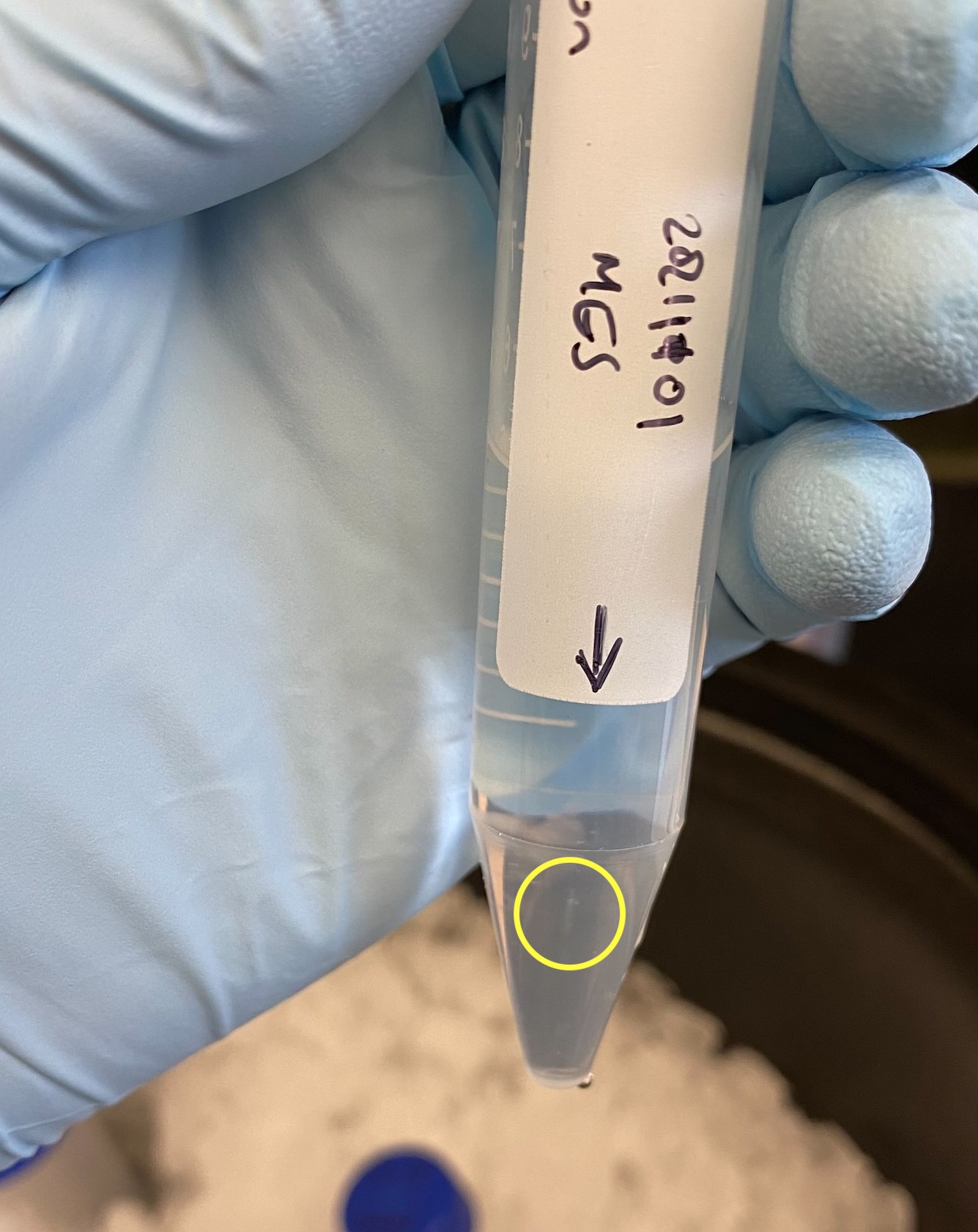
- With the fixed angle rotor, the pellet is going to be on the side of the tube. I had situated each tube in the rotor so that the side with the white patch was facing the outside of the rotor so I would at least have an idea where the pellet should be. After the first spin I thought I saw a pellet very faintly on the side of the tube (see circle in image), however the tubes had condensation and that might have cause the smudge
- Put the tubes back on ice and brought to 4055
- Removed the supernatant from each tube, pipetting from the side of the tube without the white patch to avoid any pellet
- Added 2mL cold 70% ethanol to each tube
- Vortexed tubes briefly
- Centrifuged tubes for 10 minutes at 9,500 rcf at 4 degrees C in the same centrifuge
- Looked at tubes again, I thought I saw a pellet again (although even less visible this time) but when I got back to 4055 I no longer saw one in either tube
- Tubes were transferred on ice again
- Removed the supernatant from each tube, pipetting from the side of the tube without the white patch to avoid any pellet
- Flipped the tubes upside-down on a kim wipe and let them sit for 30 minutes to dry
- After the 30 minutes, there were still some droplets in the tubes so I used a pipette tip to move them out of the tube
- When doing this, there were some droplets on the potential pellet side. Next time I tip over the tubes to let out the ethanol I need to make sure the liquid goes down the opposite side
- At this point, I could see no pellets or even smudges in the “right place”
- Added 50ul of 1X TE to each tube
- I added the droplet to the side of the tube right under where the white patch is, where I thought I saw the pellet in the image above. I then moved the tube back and forth, rotating the droplet all around the side of the tube for ~5 minutes. Then I laid the tubes on their sides with the droplets on the side of the tube and taped them down
- Tubes on their side were placed in the incubator for 1 hour at ~55 degrees C
- After incubating, the tubes were left on their sides and put on a rocking shaker overnight
20211102
- Tubes were spun down midday to collect the droplets and condensation
- I tapped off a droplet from the bottom fo the tube and rotated it around the bottom of the tube for a few minutes. This was done for each tube
- The droplets were left on the center of the lowered side of the tube and placed sideways on the rack and left on the rocking shaker for overnight again
20211103
Qubit
- Qubit reagents were allowed to come up to room temp for ~40 minutes
- Tubes were spun down to collect droplets and condensation
- The top and the bottom of the liquid lines were quantified (just in case the HMW DNA is clumped)
- Quants:
- POS2 top: 5.69 ug/mL (ng/ul)
- POS2 bottom: 6.15 ug/mL (ng/ul)
- CC2 top: 4.66 ug/mL (ng/ul)
- CC2 bottom: 4.72 ug/mL (ng/ul)
Quants this time are higher than the last successful extraction attempt. With the total volume of 50ul, the samples yielded ~200-300ng total DNA. This is actually ~10 times as much as the other attempt that worked, and I only increased the input by 2 times. This may still be low for further applications, but I think this is enough for me to do a gel on to try to separate out the 150kb size, especially if I use 15-20ul input.