Testing Zymo Quick DNA Miniprep Extraction Kit on Horseshoe Crab Tissue
DNA Extraction Test on 3 Horseshoe Crab Samples from Natalie
Samples: 463, 393, 388 Extraction kit
- Took samples out of -80 freezer and let thaw on ice bucket (took a long time)
- Made 3 1.5mL tubes labeled with sample numbers with 300ul DNA/RNA shield
- 10% bleach, DI and ETOH forceps and scalpel
- Placed new foil down for each tissue
- Took out tissue and cut a piece with new scalpel blade and tried to chop up to pre-homogenize
- Hard to cut: 463 got mostly exoskeleton I think
- 393: when trying to cut, the inside tissue squeezed out a little so no exoskeleton in piece
- 388: both exoskeleton and inside tissue
463
 388
388
 393
393

- Placed tissue pieced in 1.5mL tubes with shield
- Added 10ul of Proteinase K
- Vortexed for 10 seconds
- Placed in thermomixer 55 degrees at 1200 rpm shaking
- After 1 hour they looked the same, I added another 10ul of ProK
- After 3 hours they looked only slightly better, decided to let them digest overnight
- The next morning they looks almost the same! Even 393 that had the easiest chopped up and squishy tissue
463
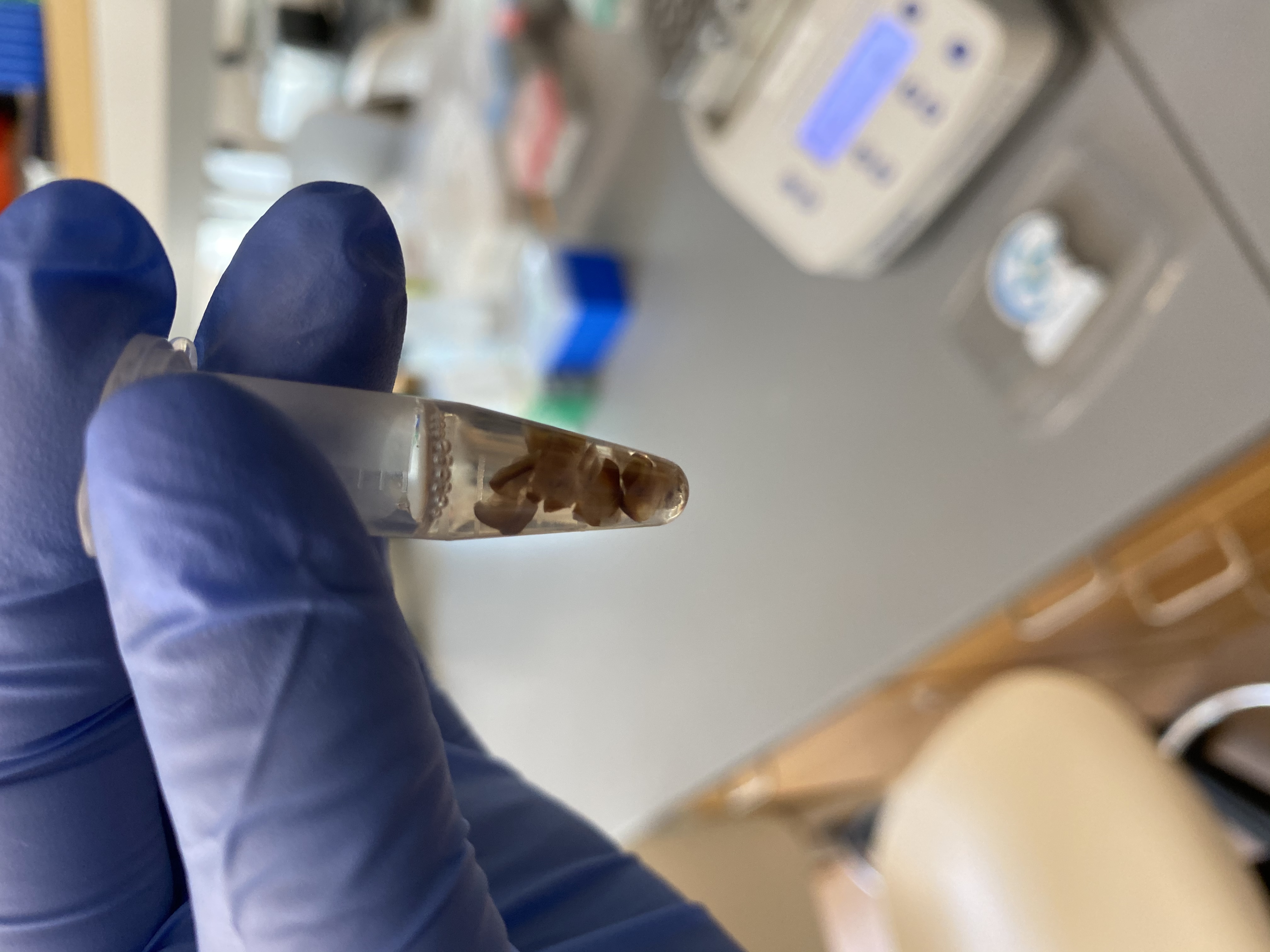 393
393
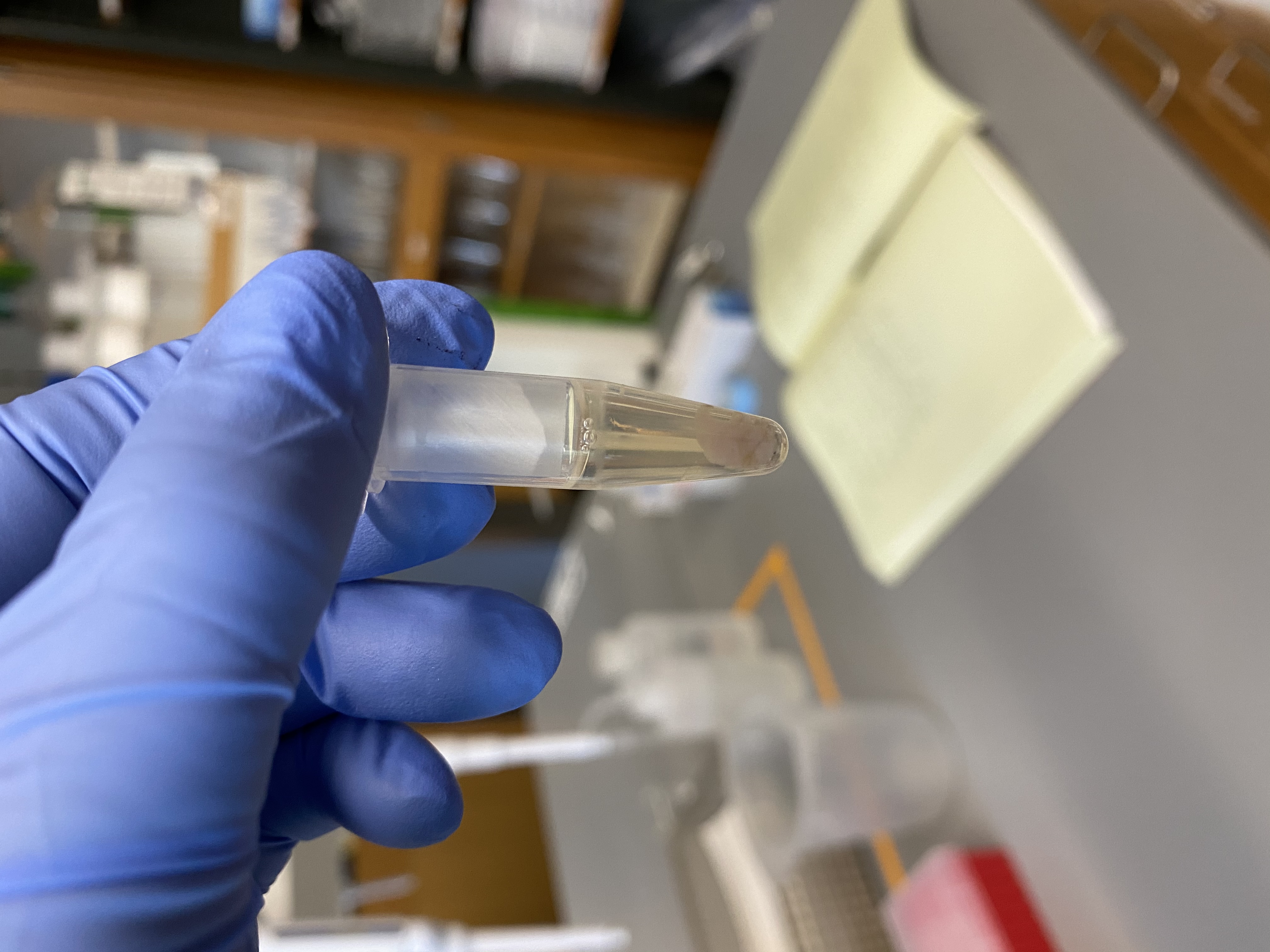
- Decided to go ahead anyways
- Centrifuged at 1300 rcf for 1 min
- Took off supernatant into new 1.5mL tubes
- There were still debris so I centrifuged the new tubes again and transferred the supernatant into new 1.5mL tubes again (~450ul)
- Added 450ul of g-DNA binding buffer to each 1.5mL tube and vortexed
- Added 700ul from each 1.5mL tube into spin columns labeled for each tube
- Centrifuged at 1300 rcf for 1 min
- Discarded flow through
- Added the rest of the liquid to the spin columns and centrifuged in the same way
- Transferred columns to new collection tubes
- Made 1.5mL tube with 10mM Tris HCl and warmed in thermomixer 55 degrees C
- Added 400ul DNA pre-wash buffer to each column
- Centrifuged 1300 rcf for 1 minute and discarded flow through
- Added 700ul DNA wash buffer to each column
- Centrifuged 1300 rcf for 1 minute and discarded flow through
- Added 400ul DNA wash buffer to each column
- Centrifuged 1300 rcf for 1 minute and discarded flow through
- Made 1.5mL tubes labeled completely with sample names and information
- Transferred spin columns to those 1.5mL tubes
- Added 50ul of warmed 10mM tris HCl by dripping to each spin column filter
- Incubated columns for 5 minutes
- Centrifuged columns for 1 min at 1300 rcf
- Repeated last 3 steps once
- Placed tubes on ice
QC
- Broad range dsDNA qubit (protocol)
- Amounts are in ng/ul and samples were read twice
| Standard 1 | Standard 2 | 463 | 393 | 388 |
|---|---|---|---|---|
| 200 | 24391 | 54.2 | 103 | 97 |
| - | - | 53.8 | 102 | 96.6 |
- Small 1% Agarose Gel (protocol)
- note that there are other samples on this gel, the last 3 are the samples extracted in this protocol
These samples have huge smearing in DNA quality. This could be because of incomplete digestion?
Written on July 27, 2020