Block 10 DNA Library Prep
DNA Library Prep for CASE EecSeq Block 10
Using the KAPA HyperPrep DNA Library Prep Kit on 18 DNA samples in 10mM Tris HCl pH 8 and 500ng from block 10 of the CASE experiment. All samples were sonicated to ~150 basepairs by using the QSonica protocol with a 27 minute time setting, 15 seconds on 15 seconds off, and an amplitude of 25%. Samples were spun down every 6 minutes.
End Repair and A-tailing
- Prepared end repair and a-tailing master mix:
- ERAT buffer 3.5μl * 19 = 66.5μl
- ERAT enzyme 1.5μl * 19 = 28.5μl
- Made 18 PCR strip tubes each with 25μl of 500ng sheared DNA
- Added 5μl of ERAT master mix to each sample
- Vortexed and spun down
- Placed samples in thermocyler A-tailing program in JONP login (~ 1 hour)
Adapter Ligation
- Prepared ligation master mix:
- ligation buffer 15μl * 19.5 = 292.5μl
- DNA ligase 5μl * 19.5 = 97.5μl
- nuclease free water 2.5μl * 19.5 = 48.7μl
- Added ligation master mix and appropriate planned adapters to each sample. Adapters were added last to minimize adapter-adapter ligation
| Sample | LMM | Adapter |
|---|---|---|
| S1 | 22.5μl | 2.5μl 1 |
| S2 | 22.5μl | 2.5μl 2 |
| S3 | 22.5μl | 2.5μl 3 |
| S4 | 22.5μl | 2.5μl 4 |
| J1 | 22.5μl | 2.5μl 5 |
| J2 | 22.5μl | 2.5μl 6 |
| J3 | 22.5μl | 2.5μl 7 |
| J4 | 22.5μl | 2.5μl 8 |
| J5 | 22.5μl | 2.5μl 9 |
| J6 | 22.5μl | 2.5μl 10 |
| J7 | 22.5μl | 2.5μl 11 |
| J8 | 22.5μl | 2.5μl 12 |
| J9 | 22.5μl | 2.5μl 1 |
| J10 | 22.5μl | 2.5μl 2 |
| J11 | 22.5μl | 2.5μl 3 |
| J12 | 22.5μl | 2.5μl 4 |
| J13 | 22.5μl | 2.5μl 5 |
| J17 | 22.5μl | 2.5μl 6 |
- Pipetted all samples up and down with the multichannel and spun down
- Incubated on the shaker at room temp for 1 hour
0.8X Cleanup
- Made fresh 80% EtOH
- Took KAPA Pure Beads out of fridge beforehand to warm to room temp
- After incubation at RT, added 44μl of KAPA pure beads to each sample and pipetted up and down at least 10 times to mix beads careful to avoid bubbles
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 12.5μl 10mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet stand and removed supernatant when clear to new labeled PCR strip tubes
Library Amplification
- Every 3 samples get a different index primer pair for amplification, so I made 6 different master mixes
- Amp MM A:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 507 primer * 3.2 = 4μl
- 1.25μl 707 primer * 3.2 = 4μl
- Amp MM B:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 508 primer * 3.2 = 4μl
- 1.25μl 708 primer * 3.2 = 4μl
- Amp MM C:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 509 primer * 3.2 = 4μl
- 1.25μl 709 primer * 3.2 = 4μl
- Amp MM D:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 510 primer * 3.2 = 4μl
- 1.25μl 710 primer * 3.2 = 4μl
- Amp MM E:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 511 primer * 3.2 = 4μl
- 1.25μl 711 primer * 3.2 = 4μl
- Amp MM F:
- 12.5μl HotStart Ready mix * 3.2 = 40μl
- 1.25μl 512 primer * 3.2 = 4μl
- 1.25μl 712 primer * 3.2 = 4μl
- Prepared new PCR tubes for the amplification with the following:
| Sample | volume adapter added DNA of sample | volume of Amp MM |
|---|---|---|
| S1 | 10μl | 15μl Amp MM A |
| S2 | 10μl | 15μl Amp MM A |
| S3 | 10μl | 15μl Amp MM A |
| S4 | 10μl | 15μl Amp MM B |
| J1 | 10μl | 15μl Amp MM B |
| J2 | 10μl | 15μl Amp MM B |
| J3 | 10μl | 15μl Amp MM C |
| J4 | 10μl | 15μl Amp MM C |
| J5 | 10μl | 15μl Amp MM C |
| J6 | 10μl | 15μl Amp MM D |
| J7 | 10μl | 15μl Amp MM D |
| J8 | 10μl | 15μl Amp MM D |
| J9 | 10μl | 15μl Amp MM E |
| J10 | 10μl | 15μl Amp MM E |
| J11 | 10μl | 15μl Amp MM E |
| J12 | 10μl | 15μl Amp MM F |
| J13 | 10μl | 15μl Amp MM F |
| J17 | 10μl | 15μl Amp MM F |
- Vortexed and spun down samples
- Placed samples in the thermocycler Genomic PCR program
1X Cleanup
- After PCR, added 25μl of KAPA pure beads to each sample and pipetted up and down at least 10 times to mix beads careful to avoid bubbles
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 16μl 10mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet stand and removed supernatant when clear to new labeled PCR strip tubes
QC
- Followed Qubit protocol for BR DNA
| Sample | Std 1 | Std 2 | Avg ng/μl |
|---|---|---|---|
| S1 | 198 RFU | 21757 RFU | 67 |
| S2 | - | - | 150 |
| S3 | - | - | 163 |
| S4 | - | - | 134.5 |
| J1 | - | - | 171.5 |
| J2 | - | - | 115.5 |
| J3 | - | - | 165.5 |
| J4 | - | - | 138.5 |
| J5 | - | - | 163 |
| J6 | - | - | 119.5 |
| J7 | - | - | 146 |
| J8 | - | - | 150.5 |
| J9 | - | - | 169.5 |
| J10 | - | - | 162.5 |
| J11 | - | - | 165 |
| J12 | - | - | 141.5 |
| J13 | - | - | 166.5 |
| J17 | - | - | 143 |
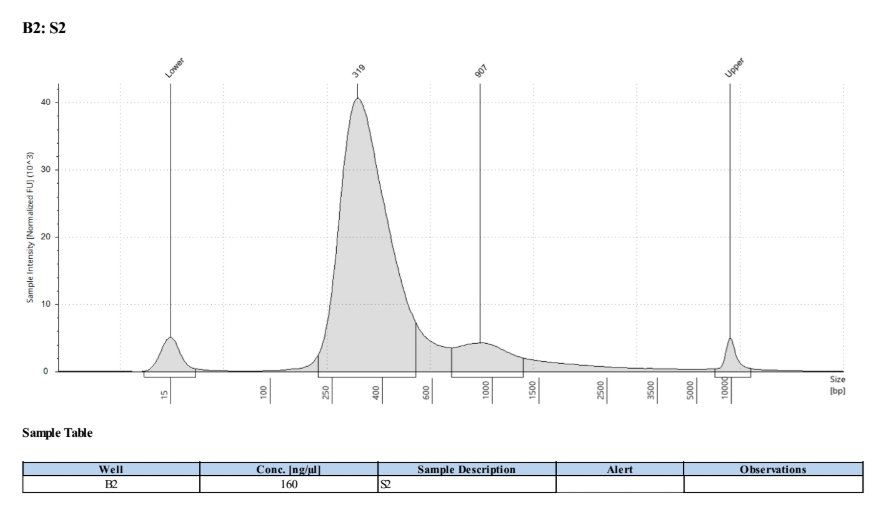
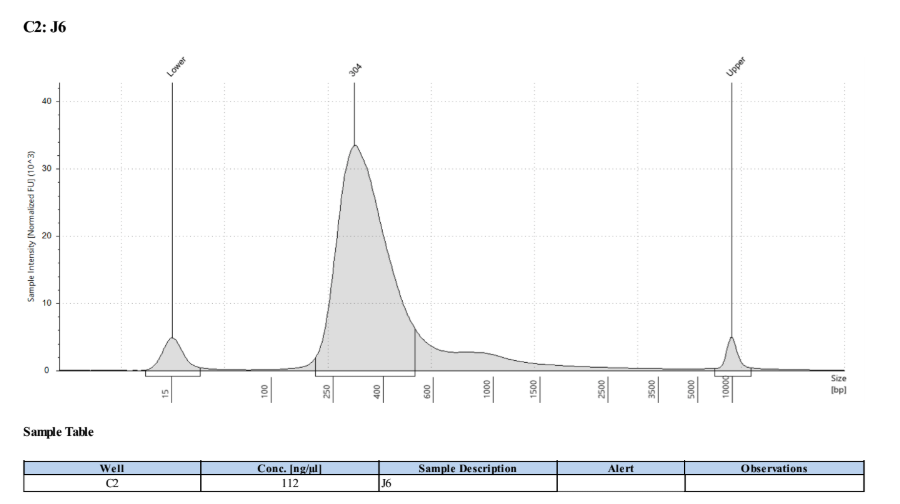
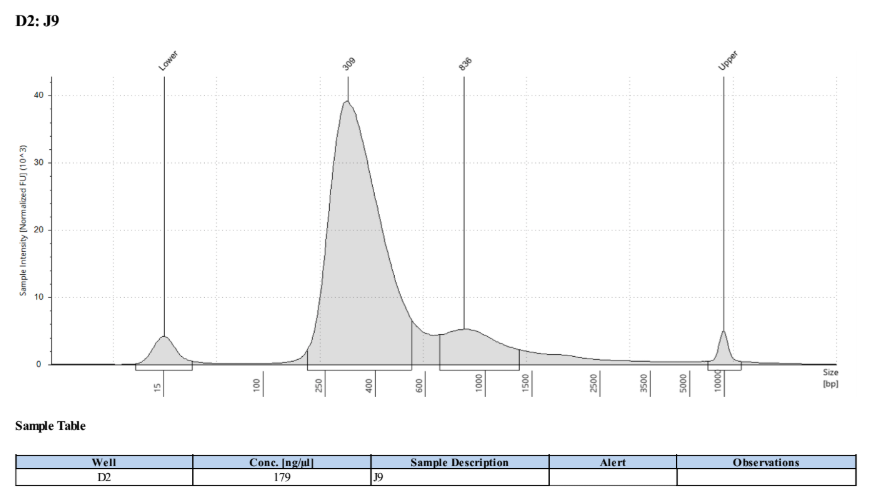
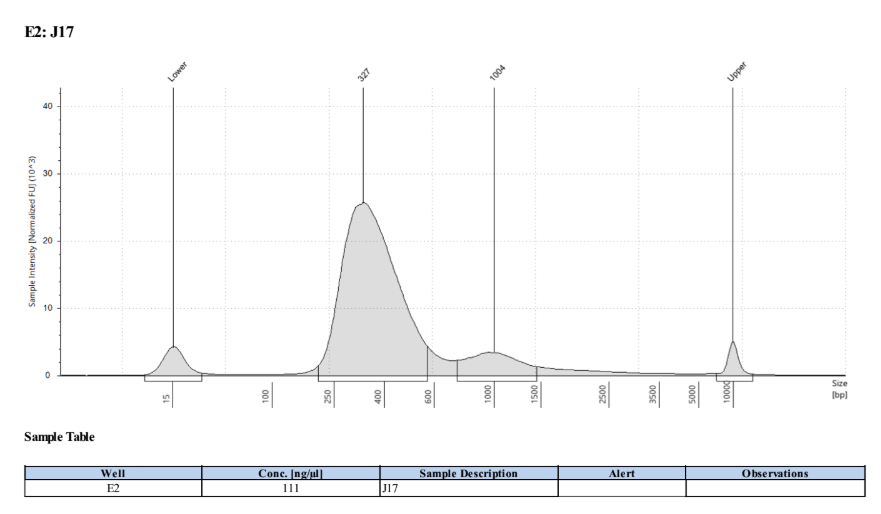
- Followed tapestation protocol for D5000 tapes on 4 representative samples to check
See full report here
Written on September 25, 2019