CASE Block 12 Probe Synthesis and Hybridization
Synthesis of EecSeq Probes from Block 12 cDNA Libraries and Hybridization of Probes to DNA Libraries
Normalization
Pooling of cDNA libraries, 200ng each
| Sample | μl cDNA |
|---|---|
| B12T24J1 | 1.68 |
| B12T24J2 | 1.60 |
| B12T24J3 | 1.72 |
| B12T24J4 | 1.85 |
| B12T24J5 | 1.96 |
| B12T24J6 | 2.85 |
| B12T24J7 | 1.88 |
| B12T24J8 | 1.58 |
| B12T24J9 | 1.56 |
| B12T24J10 | 7.33 |
| B12T24J11 | 2.44 |
| B12T24J12 | 5.48 |
| B12T24J13 | 2.02 |
| B12T24J14 | 2.54 |
| B12T24J15 | 2.01 |
| B12T24J18 | 2.04 |
Total Volume: 40.53μl
BR Qubit quant of pool: 82.8ng/μl
DSN Treatment
- 500ng of pool used for DSN treatment: 6.04μl in a new PCR tube
- Brought up to 13.5μl with 7.96μl nuclease-free H20
- 4X Hybridization buffer made
- 4.5μl of 4X hybridization buffer added to the tube
- Thermocycler DSN program
- Diluted 10X DSN Master Buffer to 2X
- Put diluted master buffer in 2nd thermocyler at 68 degree hold for 10 min to come up to temp
- Added whole volume of 2X DSN master buffer to the pooled sample in the first thermocycler, pipetted to mix inside the thermocycler
- Waited 10 minutes at 68 degrees C
- Added 2μl of DSN enzyme to pooled sample in first thermocycler, pipetted to mix with p200 pipette
- Waited 25 minutes at 68 degrees C
- Added 40μl DSN stop solution to the tube and pipetted to mix, placed on ice
Post-DSN
- Did 1.6X clean up: 128μl KAPA Pure Beads
- Eluted in 25μl 10mM Tris HCl
- High Sensitivity Qubit: 1.11ng/μl
Post-DSN PCR
- 20μl DSN treated library
- 25μl KAPA ready mix
- 5μl KAPA library amp primer mix
- Placed in thermocycler 14 cycle DSN PCR program
- After: 1.6X cleanup: 80μl KAPA Pure Beads
- Eluted in 22μl 10mM Tris HCl
- Broad Range Qubit: 5.45ng/ul
Seems like PCR didn’t work great, but this has happened before. Wanted to check the tapestation to make sure nothing is weird
D5000 TapeStation
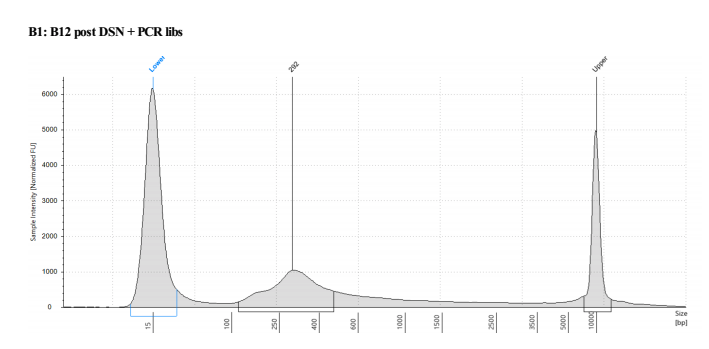
Looks mostly right, except there is primer dimer in there so I decided to do another bead cleanup before splitting it into 4 replicate tubes and doing 8 cycle PCRs on those
Cleanup and PCR
- 1.6X cleanup so 32μl KAPA Pure Beads
- Eluted in 24μl 10mM Tris HCl
- Set up 4 PCR tubes with 5μl pooled-treated libs in each, saving 4μl if necessary
- PCR master mix:
- 52.5μl KAPA ready mix
- 10.5μl KAPA lib amp primers
- 21μl nuclease free water
- Thermocycler 8 cycle DSN PCR program
- Pool together 4 tubes after (100μl) and 1.6X cleanup: 160μl KAPA Pure Beads
- Eluted in 40μl 10mM Tris HCl
- Broad Range Qubit: 132.5ng/μl!!
Probe Synthesis
Saving 1μg of probes and doing 4 separate digestions each with 1μg. There is enough for that.
Restriction Digest of Adapters and MBN Treatment
- Made 4 PCR tubes with 7.55μl of pooled-treated libraries (1μg each)
- Brought up to 12.25μl with 7.7μl nuclease free water
- Made digest master mix:
- 16.4μl 10X cutsmart buffer
- 4.1μl SAII enzyme
- 93.28μl nuclease free water
- Added 27.7μl of mastermix to each PCR tube on ice
- Theremocycler RE digest short program 4 hours
- Made Mungbean mastermix
- 18.9μl 10X mungbean buffer
- 2.1μl mungbean nuclease enzyme
- Added 5μl of mastermix to each tube in thermocycler then did MBN program 30 min
- Did a 1.5X bead cleanup on each tube: 67.5μl KAPA Pure Beads
- eluted in 21μl 10mM Tris HCl
- Did another 1.5X bead cleanup on each tube: 31.5μl KAPA Pure Beads
- Eluted in 22μl 10mM Tris HCl
D5000 Tapestation results look right, no left over adapters, and the size has shifted smaller with the loss of the adapters
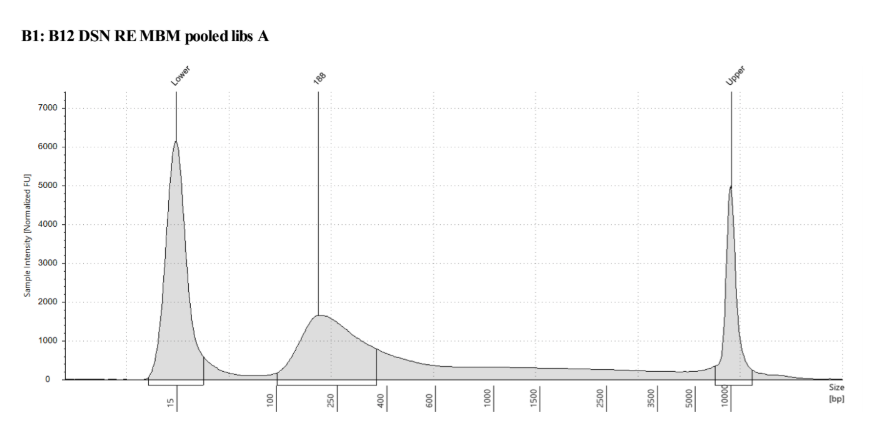
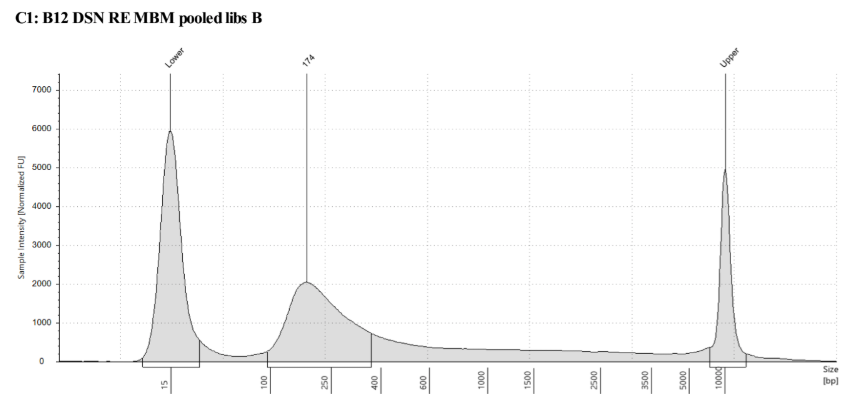
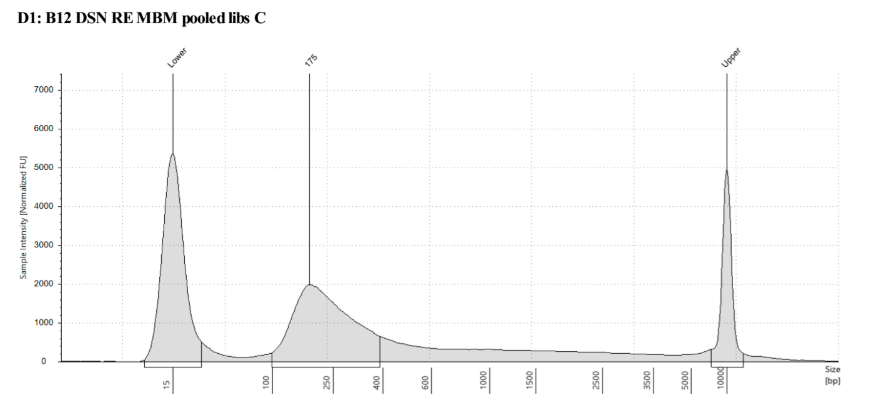
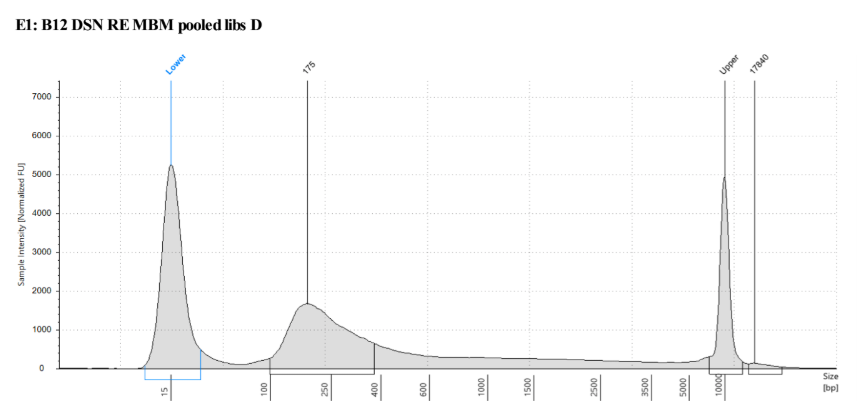
Biotin Labeling
- Made biotin mastermix:
- 42μl 5X decanucleotide reaction buffer
- 58.8μl nuclease free water
- Added 24μl of mix to 4 PCR tubes
- Added 20μl of DSN-RE-MBN treated pooled libraries, one for each tube
- Put in thermocycler Biotin program, ~10 min until a 4 degree hold
- Make biotin labeling mastermix:
- 21μl biotin labeling mix
- 4.2 klenow fragment
- Added 6μl of biotin labeling master mix to each of the 4 tubes
- Shook tubes and spun down, continued program ~20 hours
- Pooled all samples together for 1.5X cleanup: 300μl KAPA pure beads
- Eluted in 50μl 10mM Tris HCl pH 8
- Qubit: 92.8ng/μl!!
D5000 Tapestation results look normal to me! Still not sure why it trails off so much
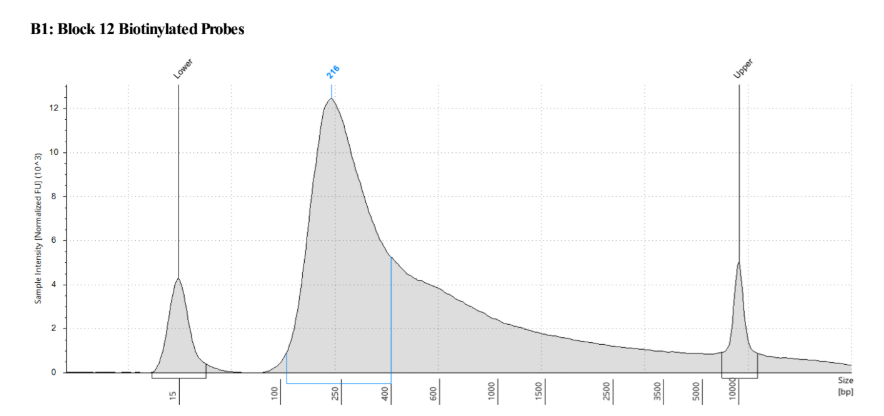
A small detour
Note, first attempt of hybridization, I failed to add the probes to the capture pools… thus it needed to be done over again and one sample need to have more genomic DNA library created. I also tried to recover the capture pools from the starting wash of the capture.
B12T0S3 gDNA lib prep
- End repair and A-tailing
- 25μl sheared DNA sample
- 3.5μl ERAT buffer
- 1.5μl ERAT enzyme
- Thermocyler ERAT program ~1hr
- Adapter ligation
- T0S3 gets adapter #3
- 15μl ligation buffer
- 2.5μl nuclease free water
- 5μl ligase enzyme
- 2.5μl adapter #3, added after the 30μl of end-repaired sample is added
- Incubated on the shaker at room temp for 1 hour
- 0.8X cleanup
- Beads to room temp and new 80% EtOH
- 44μl KAPA pure beads added to ligation mix with sample
- Normal bead cleanup performed
- Resuspended and retained sample in 11.5μl 10mM tris HCl pH 8
- Index addition
- T0S3 gets index pair 509-709, reaction set up on ice
- 10μl of adapter ligated sample used for PCR
- 12.5μl KAPA ready mix
- 1.25μl 509 index
- 1.25μl 709 index
- Genomic PCR program in thermocycler
- 1X cleanup
- 25μl of KAPA pure beads
- Resuspended and retained sample in 16μl of 10mM Tris HCl pH 8
- 1.5X cleanup of starting supernatant from 1st capture “attempt”
- Volume in in tubes is ~223μl
- 1.5X is 334.5μl KAPA pure beads
- Performed normal bead cleanup
- Resuspended and retained samples in 16μl of 10mM Tris HCl pH 8
- Broad range Qubit of above
| standard 1 | standard 2 | T0S3 | recovery capture 1 | recovery capture 2 | recovery capture 3 | recovery capture 4 |
|---|---|---|---|---|---|---|
| 189 RFU | 21209 RFU | 18.7ng/μl | 36.3ng/μl | 36.9ng/μl | 38.1ng/μl | 40ng/μl |
The DNA library doesn’t have a great concentration, but the size is right and it is enough for the capture so I went with it
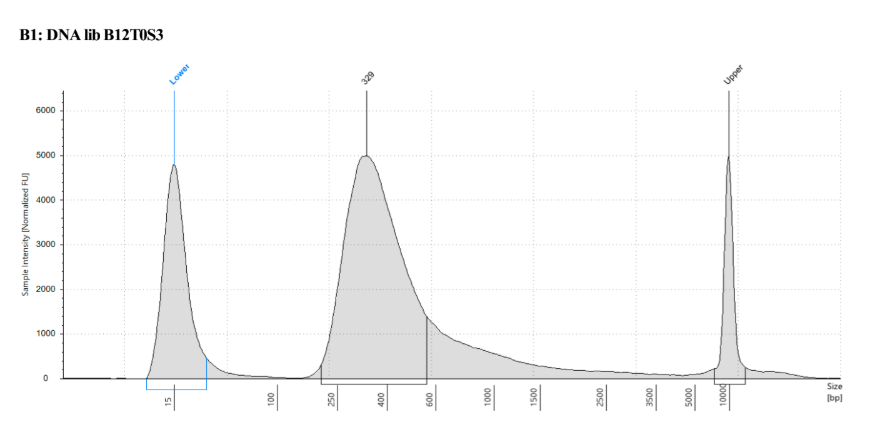
Prep and Hybridization
Pooling of DNA Libraries
- Pool 200ng of each DNA library, add 500ng of probes, then add water to equal up to 23.5μl total volume
- Probes quant is 92.8ng/μl, so 500ng is 5.39μl
Capture 1
| Sample | vol DNA for 200ng |
|---|---|
| T0S1 | 6.17μl |
| T24J1 | 1.59μl |
| T24J2 | 2.16μl |
| T24J3 | 1.5μl |
| T24J5 | 1.72μl |
| Total | 13.14μl |
Capture 2
| Sample | vol DNA for 200ng |
|---|---|
| T0S2 | 5μl |
| T24J4 | 1.61μl |
| T24J6 | 1.49μl |
| 24J7 | 1.72μl |
| T24J8 | 2μl |
| Total | 11.82μl |
Capture 3
| Sample | vol DNA for 200ng |
|---|---|
| T0S3 | 10.9μl |
| T24J9 | 1.6μl |
| T24J10 | 1.51μl |
| T24J11 | 1.89μl |
| T24J13 | 1.9μl |
| Total | 17.8μl |
Capture 4
| Sample | vol DNA for 200ng |
|---|---|
| T0S4 | 6.78μl |
| T24J12 | 1.7μl |
| T24J14 | 2.25μl |
| T24J15 | 2.04μl |
| T24J18 | 1.86μl |
| Total | 14.63μl |
- 5.39μl of block 12 probes were added to each capture pool
- The volume of nuclease free water to add up to 23.5μl total in each capture pool was calculated and added:
| Sample | vol pool w/probes | vol H20 |
|---|---|---|
| Cap 1 | 18.53μl | 4.97μl |
| Cap 2 | 17.22μl | 6.28μl |
| Cap 3 | 23.7μl | NA |
| Cap 4 | 20.02μl | 3.48μl |
Hybridization
- Made hybridization master mix (volumes for 1 hybridization multiplied by 4.3):
- 51.6μl 20X SSC
- 1.72μl 500mM EDTA
- 1.72μl 10% SDS
- 6.88μl 50X Denhardt’s solution
- 2.15μl human COT-1 DNA
- 1.72μl blocking oligo 1
- 1.72μl blocking oligo 2
- 1.72μl blocking oligo 3
-
1.72μl blocking oligo 4
- Added 16.5μl of hybridization mix to each capture pool and pipetted to mix note the mix had a white precipitate that mostly vortexed away, but I tried to keep it evenly distributed to each capture
- Put in the thermocycler hybridization program 95C for 10 minutes then moved to the already warmed incubator genie 65C rocking 10 speed for 48 hours, I tried to parafilm the lids to protect from evaporation. Samples were taken out and vortexed and spun down once
Washes and Solutions for Capture
TEN
- 10mM Trish HCl pH 7.5, 1mM EDTA, 1M NaCl
- Need 3,600μl
- 7.2μl of 500mM EDTA
- 720μl of 5M NaCl
- 2872.3μl of 10mM Tris HCl pH 7.5 (used the Tris we made last time, checked the pH and it was 7.51)
Solution 1
- 1X SSC, 0.1% SDS
- Warmed to 65C in thermomixer, put into two 1.5mL tubes with 850μl in each
- Need 1700μl (used twice)
- 85μl 20X SSC
- 17μl 10% SDS
- 1598μl nuclease free H20
Solution 2
- 0.5X SSC, 0.1% SDS
- Need 900μl
- 22.5μl 20X SSC
- 9μl 10% SDS
- 868.5μl nuclease free H2O
Solution 3
- 0.1X SSC, 0.1% SDS
- Need 900μl
- 4.5μl 20X SSC
- 9μl 10% SDS
- 886.5μl nuclease free H2O
Prepare Beads
- Resuspended Dynabeads M-280 beads first with p200
- Made 4 PCR tubes with 10μl of resuspended beads in each
- Added 200μl of prepared TEN solution to each tube and pipetted to mix
- Placed tubes on magnet plate and removed supernatant when clear
- Removed from magnet and resuspended beads in 200μl TEN solution
- Repeated wash twice for a total of 3 washes
- Resuspended beads in 200μl TEN solution
Capture
- After the ~48 hour hybridization incubation, took the hybridized pools out of the incubator, spun them down, and added the 4 hybridization mixtures to each of the 4 washed and prepared Dynabead PCR tubes note: volume in hybridization tubes was closer to 37μl, potentially there was evaporation
- Pipetted to mix and incubated on the shaker at room temp for 30 minutes
- Placed solution 1 in thermomixer to get to 65°C
- Turned on thermocycler and set it on the 65°C hold program
-
Made PCR strip tubes to save every wash (20 total tubes)
- Placed tubes on magnet plate after incubation
- Removed supernatant when clear and saved as “start”
- Resuspended beads in 200μl 65°C solution 1
-
Placed tubes in thermocyler set at 65°C for 15 minutes
- Placed tubes on magnet plate after incubation
- Removed supernatant when clear and saved as “wash 1”
- Resuspended beads in 200μl 65°C solution 1
-
Placed tubes in thermocyler set at 65°C for 10 minutes
- Placed tubes on magnet plate after incubation
- Removed supernatant when clear and saved as “wash 2”
- Resuspended beads in 200μl room temp solution 2
-
Placed tubes in thermocyler set at 65°C for 10 minutes
- Placed tubes on magnet plate after incubation
- Removed supernatant when clear, note: forgot to save wash at this step :(
- Resuspended beads in 200μl room temp solution 3
-
Placed tubes in thermocyler set at 65°C for 10 minutes
- Turned on second thermocycler and set it on the 80°C hold program
-
Placed 200μl of nuclease free H20 in the 80°C thermocycler
- Placed tubes on magnet plate after incubation
- Removed supernatant when clear and saved as “wash 4”
- Resuspended beads in 22μl 80°C nuclease free water
-
Placed tubes in thermocyler set at 80°C for 10 minutes
- Made PCR master mix for amplification:
- 12.5μl KAPA ready mix * 4.5 = 52.5μl
- 2.5μl KAPA universal primer mix * 4.5 = 10.5μl
-
Aliquoted 15μl of PCR master mix into each of 4 PCR tubes labeled for each capture
- Placed Dynabead tubes on magnet plate after incubation
- SAVED supernatant when clear as captured DNA into new PCR tubes
- 10μl of captured DNA was transferred to their corresponding PCR tubes containing the amplification master mix and were vortexed and spun down
- Tubes for amplification were placed in the thermocyler for the post-capture 12-cycle PCR program
- The other tubes (all washes and the remaining ~10μl of captured DNA) were placed in the -20 freezer
- After the PCR, a 1X bead cleanup was performed:
- Added 25μl of KAPA pure beads to each sample
- Performed normal bead cleanup
- Resuspended and retained captured DNA in 25μl 10mM Tris HCl
High Sensitivity Qubit:
| standard 1 | standard 2 | Capture 1 post-PCR | Capture 2 post-PCR | Capture 3 post-PCR | Capture 4 post-PCR |
|---|---|---|---|---|---|
| 38.94 RFU | 26483 RFU | 17.9ng/μl | 18.5ng/μl | 19.4ng/μl | 13.2ng/μl |
D5000 TapeStation to check that the right stuff was in the sample:
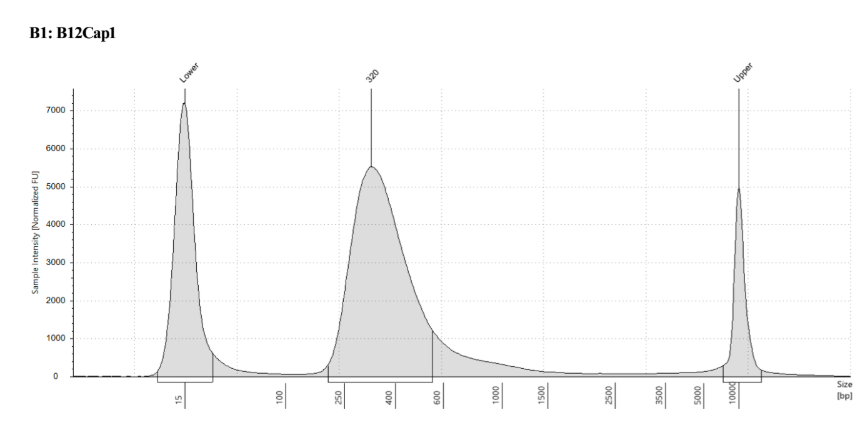
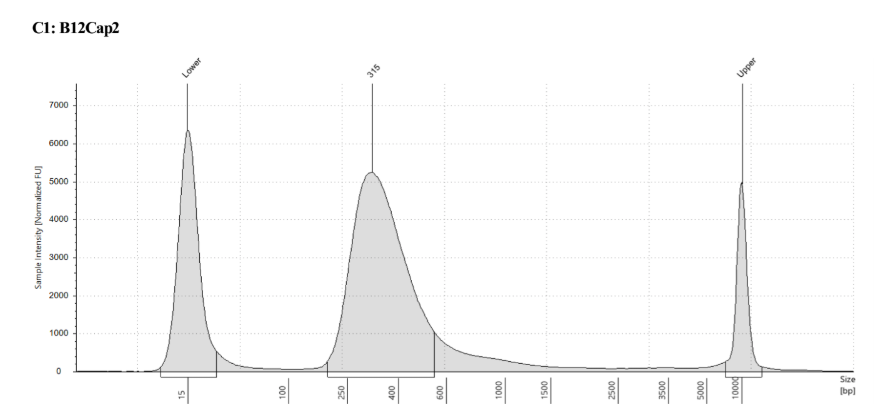
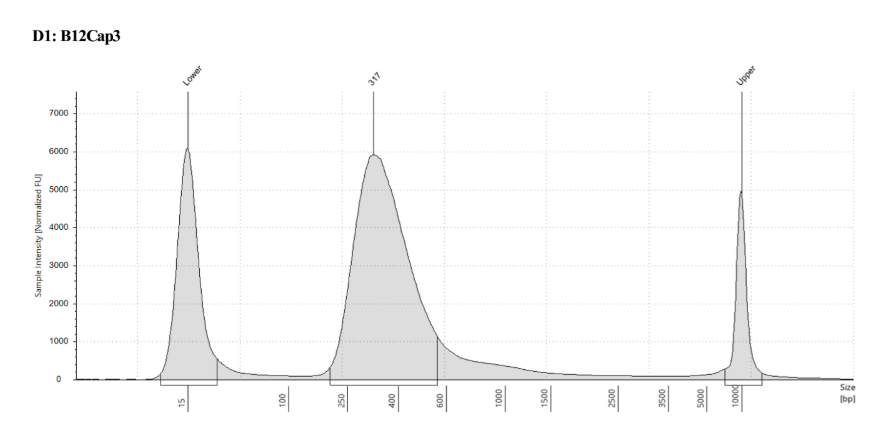
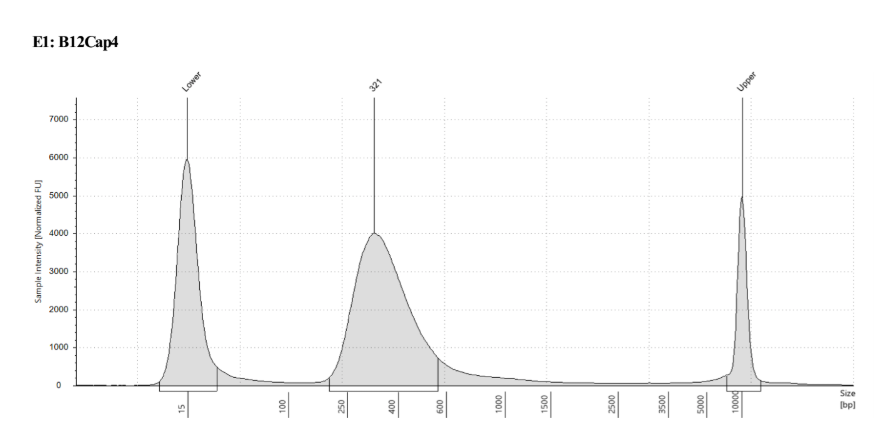
full results