Australia Mytilus EecSeq Probe Synthesis Day 6
Enrichment of DSN Treated Libraries
Continuing with Amy Zyck and also Jacob Green
- Take tubes out of the freezer and thaw
- Warm KAPA pure beads to room temp and make new 80% EtOH
- Added 128μl KAPA pure beads to each of the two tubes and pipetted to mix
- Placed on shaker for 15 minutes
- Placed tubes on magnet plate and removed the supernatant when clear
- Added 200μl 80% EtOH to each tube carefully
- Removed ~180μl supernatant from each tube carefully
- Added 200μl 80% EtOH to each tube carefully
- Removed ALL supernatant from each tube and used p20 tips to remove any EtOH droplets
- Resuspended beads in 25μl 10mM Tris HCl pH 8
- Put tubes on shaker for 5 minutes
- Placed tubes on magnet stand and removed 25μl of clear supernatant into new tubes
- Performed HS Qubit on DSN treated libraries (Followed this protocol)
- S1: 64 RFU
- S2: 25850 RFU
- Tube 1: 0.718ng/μl
- Tube 2: 0.762ng/μl
PCR Enrichment of DSN Treated Libraries
- Master mix for PCR:
- 25μl 2X KAPA HiFi Hot Start ready mix * 2.2 = 55μl
- 5μl 10X KAPA library amplification primer mix * 2.2 = 11μl
- Made 2 new tubes for the PCR
- Added 30μl master mix each to the new tubes
- Added 20μl of DSN treated library pools to the appropriate tube for a total volume of 50μl
- Votexed to mix and spun down
- Placed in the thermocycler DSN PCR program 14 cycles
- Took KAPA beads out of the fridge to warm to room temp
- Afterwards, spun down and a 1.6X bead clean up
- Added 80μl KAPA pure beads to each tube and pipetted to mix
- Placed on shaker for 15 minutes
- Placed tubes on magnet plate and removed the supernatant when clear
- Added 200μl 80% EtOH to each tube carefully
- Removed ~180μl supernatant from each tube carefully
- Added 200μl 80% EtOH to each tube carefully
- Removed ALL supernatant from each tube and used p20 tips to remove any EtOH droplets
- Resuspended beads in 22μl 10mM Tris HCl pH 8
- Put tubes on shaker for 5 minutes
- Placed tubes on magnet stand and removed 22μl of clear supernatant into new tubes
- Performed BR Qubit on DSN treated libraries (Followed this protocol)
- S1: 197 RFU
- S2: 21088 RFU
- Tube 1: 2.5ng/μl
- Tube 2: 7.2ng/μl
- Ran D5000 TapeStation on the two post DSN normalized samples
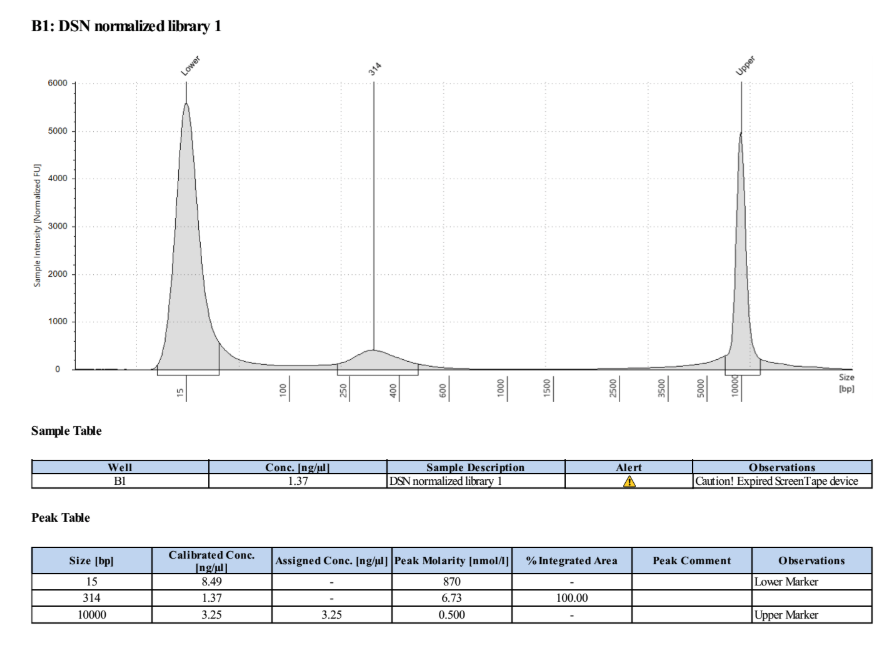
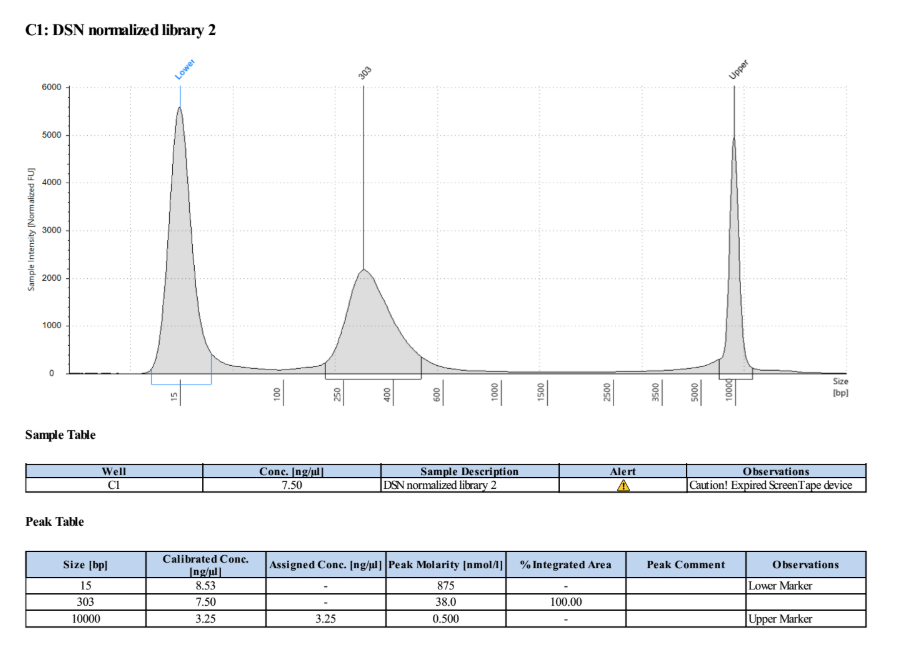
It’s really hard to tell, especially in the first sample, if there is any primer or left over digested adapter in the traces because the amount is so low. We did a second PCR of the already PCR’d samples to check - Took 2μl from each of the already PCR’d samples and added 18μl of 10mM Tris HCl pH 8 to each to get to 20μl.
- Made the same master mix as above, 55μl KAPA ready mix and 11μl of KAPA primer mix
- 30μl master mix into the new tubes with the diluted 2μl of PCR’d samples
- Put in the thermocycler same program DSN PCR 14 cycles
- Performed BR Qubit (Followed this protocol)
- S1: 193 RFU
- S2: 19922 RFU
- Tube 1: 223ng/μl
- Tube 2: 252ng/μl
- Ran the TapeStation (D5000) to check for smaller fragments now that the concentration is so high
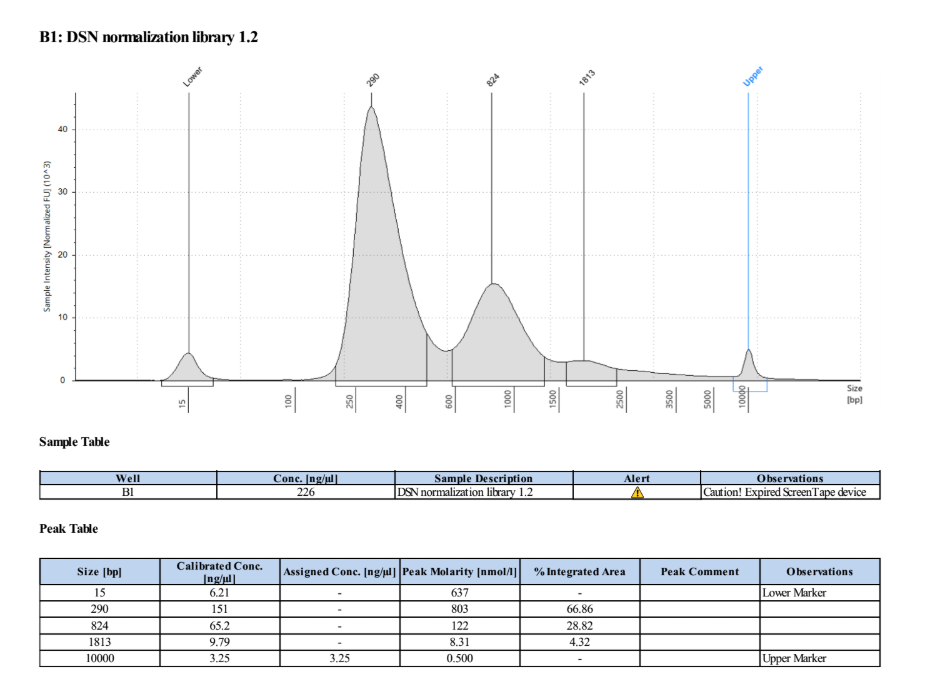
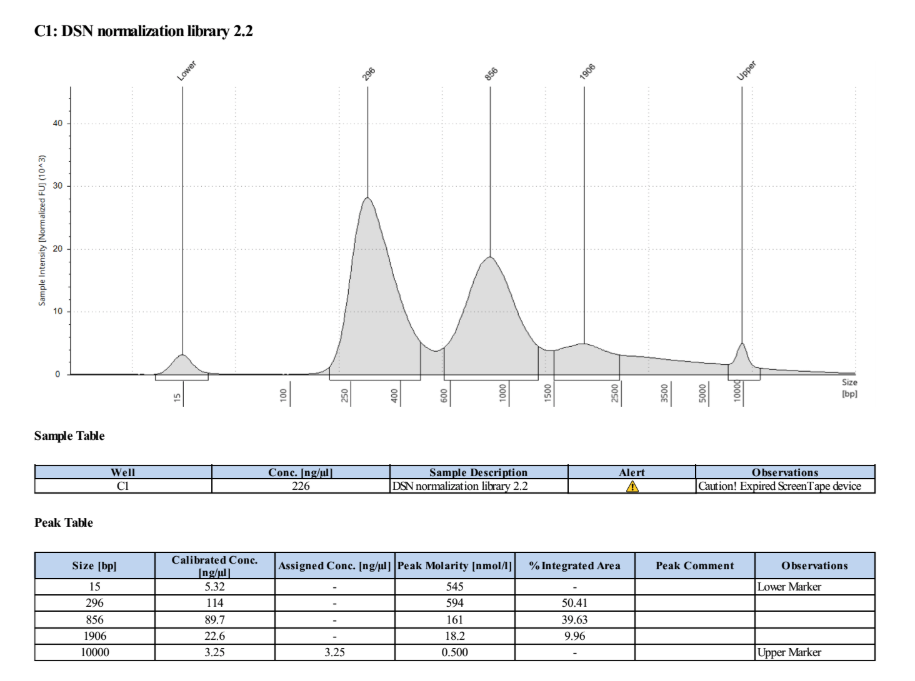 The peaks above ~300bp are evidence of overamplification and secondary structures of DNA in the reactions, but there is no amplification of adapter or primers in these samples which is the important part.
The peaks above ~300bp are evidence of overamplification and secondary structures of DNA in the reactions, but there is no amplification of adapter or primers in these samples which is the important part. - To proceed, we did another 8 cycle amplification from the original post DSN PCR’d samples (see step 32). To avoid overamplification, three reactions with 5μl each per sample were made for a total of 6 reactions
- Made master mix for the PCR:
- 12.5μl KAPA ready mix * 6.2 = 77.5μl
- 2.5μl KAPA primer mix * 6.2 = 15.5μl
- Added 5μl of sample 1 DNA to 3 new PCR tubes, and added 5μl of sample 2 DNA to 3 new PCR tubes
- Added 5μl of 10mM Tris HCl to each of those to dilute to 10μl
- Added 15μl of the amplification master mix to each tube for a total reaction volume of 25μl
- Vortexted, spun down, and placed in the thermocycler DSN PCR program 8 cycles
- Took KAPA pure beads out of the 4 degree to warm to room temp
- After the PCR was finished, pooled the 3 tubes from each set together for 2 tubes with 75μl
- Performed a 1.6X cleanup by adding 120μl KAPA beads and pipetting to mix
- Placed on shaker for 15 minutes
- Placed tubes on magnet plate and removed the supernatant when clear
- Added 200μl 80% EtOH to each tube carefully
- Removed ~180μl supernatant from each tube carefully
- Added 200μl 80% EtOH to each tube carefully
- Removed ALL supernatant from each tube and used p20 tips to remove any EtOH droplets
- Resuspended beads in 22μl 10mM Tris HCl pH 8
- Put tubes on shaker for 5 minutes
- Placed tubes on magnet stand and removed 22μl of clear supernatant into new tubes
- Performed BR Qubit (Followed this protocol)
- S1: 191 RFU
- S2: 20665 RFU
- Tube 1: 138ng/μl
- Tube 2: 196ng/μl
- Ran the TapeStation (D5000) to check for overamplification and the right distribution
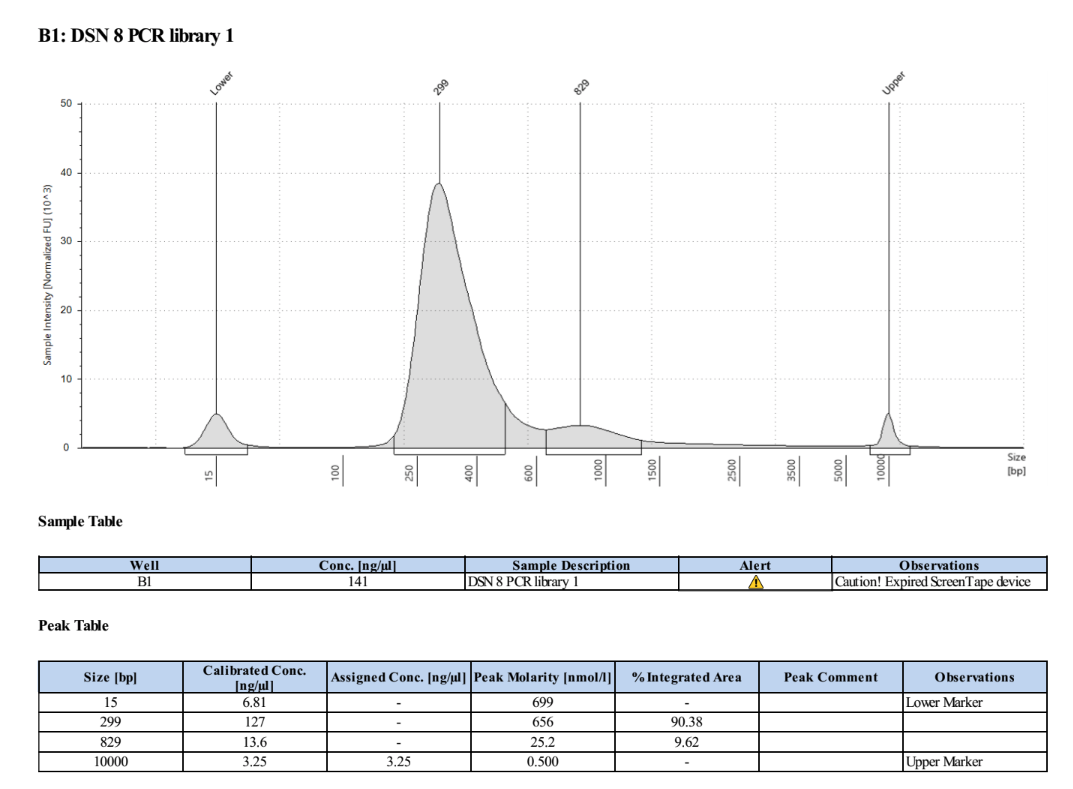
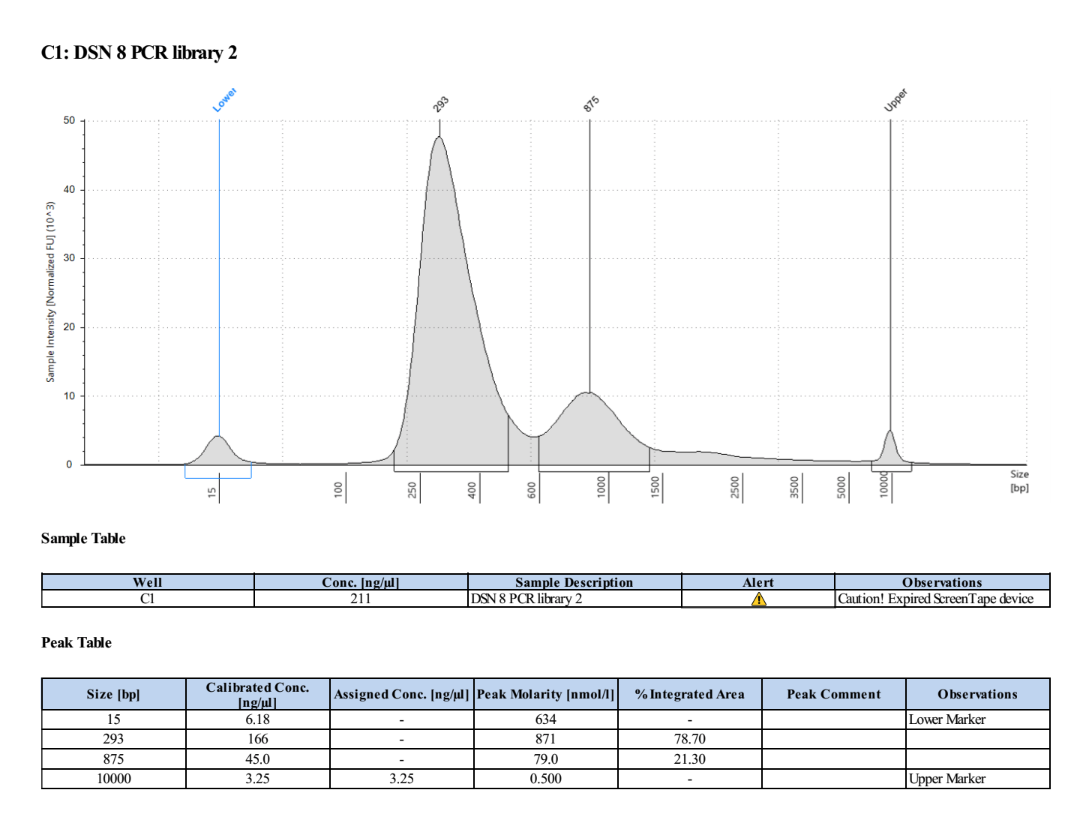 There is a lot more DNA in these and much less overamplification, success!
There is a lot more DNA in these and much less overamplification, success!
Tubes placed in the fridge overnight
Written on June 28, 2019